NegMiner: An Automated Tool for Mining Negations from Electronic Narrative Medical Documents
Автор: Hanan Elazhary
Журнал: International Journal of Intelligent Systems and Applications(IJISA) @ijisa
Статья в выпуске: 4 vol.9, 2017 года.
Бесплатный доступ
Mining negations from electronic narrative medical documents is one of the prominent data mining applications. Since medical documents are freely written, it is impossible to consider all possible sentence structures in advance and so frequent update of mining algorithms is inevitable. Unfortunately most of the proposed algorithms in the literature are too complex to be easily updated. Besides, most of them cannot be easily ported to other natural languages. The simple NegEx algorithm utilizes only two regular expressions and sets of terms to mine negations from narrative medical documents and so does not suffer from these shortcomings. Meanwhile, it has shown impressive mining results and so it is the most widely adopted algorithm. This paper proposes the Negation Mining (NegMiner) tool to address some of the shortcomings of the NegEx algorithm. The NegMiner exploits some basic syntactic and semantic information to deal with contiguous and multiple negations. It is a user-friendly tool that facilitates the task of knowledge base update and the task of document analysis through the use of PDF files. This also makes it able to deal with the existence of a medical finding several times in a single sentence. Experimental results have shown the superiority of the mining results of the NegMiner in comparison to the simulated NegEx algorithm.
Data Mining, Medical Documents, Natural Language Processing, Negations, NegEx, NLP
Короткий адрес: https://sciup.org/15010917
IDR: 15010917
Текст научной статьи NegMiner: An Automated Tool for Mining Negations from Electronic Narrative Medical Documents
Published Online April 2017 in MECS
Narrative medical documents including radiology and clinical examination reports, discharge summaries and prescriptions can provide a wealth of information in order to support making decisions regarding the patients, making predictions about diseases, identifying patients eligible for specific research studies, indexing and for research purposes. Unfortunately, most of the findings and diseases in such documents are negated and information retrieval techniques typically do not differentiate between absent and present findings [1] in a document. This led to the development of specialized algorithms for mining negations from electronic narrative medical documents.
Negation mining algorithms are generally based on the assumption that narrative medical documents are formed of sentences and that each sentence includes negation phrases or cues1 (such as without and denies ) in addition to Unified Medical Language System (UMLS) terms. The UMLS [2] provides unified terminology, classification and coding standards for compatible Electronic Health Records (EHRs) and biomedical information systems. Each finding or disease in a narrative medical document is assumed to be a UMLS term with a unique UMLS string ID [1]. The goal of a negation mining algorithm is to determine the UMLS terms that are negated by negation phrases and cues in the corresponding sentences.
Many research studies aimed at exploring possible negation phrases and negations and/or preparing sets of sentences for negation mining research. For example, Chapman et al. [3] developed a lexicon of possible negation phrases and cues in multiple languages and prepared a set of test sentences for research purposes [4]. Morante [5] enumerated negation cues and their scopes in biomedical documents. Huang and Lowe [6] provided a grammar-based classification of negations through the analysis of 500 radiology reports. The free BioScope corpus [7] includes sentences annotated with information about the scope of negation cues.
The problem of mining negations from electronic narrative medical documents has been tackled in several research studies in the literature. Algorithms for negation mining can be classified broadly into rule-based algorithms, machine leaning algorithms and hybrid algorithms. Rule-based algorithms generally rely on the use of regular expressions in addition to syntactic and semantic information to infer the scope of the negation cues and thus determine the negated UMLS terms. Alternatively, machine learning algorithms, as the name implies, rely on machine learning methods and classification techniques for this purpose. The simplest and most widely employed algorithm is the rule-based NegEx algorithm [1] that relies on two regular expressions in addition to a set of negation and termination terms to determine the negated UMLS terms. In spite of the simplicity of this algorithm, it has shown impressive mining results and so it has been ported from English to several other natural languages [8-9]. Although some other rule-based and machine learning algorithms have reported slightly better results, they have not been widely adopted due to their complexity and the difficulty of porting them to other natural languages. For example, most rule-based algorithms rely on syntactic and semantic information and so are natural languagedependent. Besides, since medical documents are written by humans, we can never consider every possible sentence structure in advance and hence both machine learning algorithms and rule-based algorithms need to be frequently updated. The more complex an algorithm is, the harder and more time-consuming is the update task.
In summary, the NegEx is favored over other rulebased and machine learning algorithms in the literature due to several reasons. First, in spite of its simplicity, this algorithm has shown satisfactory results. Second, it can be easily translated and adapted to other natural languages and can be easily updated. Besides, the other algorithms in the literature have not shown considerable improvement in performance in spite of their complexity. Additionally, medical text is more restricted than free text and thus does not require formal natural language processing (NLP) [1]. This algorithm is, thus, the most widely adopted algorithm. It is still being exploited in many applications and is being ported to many natural languages as explained in the following section. Accordingly, we argue that more effort need to be exerted to improve the NegEx algorithm rather than switching to a more complex algorithm.
This paper proposes the Negation Mining (NegMiner) tool to address some of the shortcomings of the NegEx algorithm. It exploits basic syntactic and semantic information to deal with contiguous and multiple negations, unlike the NegEx algorithm. Additionally, the NegMiner is a user-friendly tool that facilitates the process of knowledge base update and document analysis. It also generates explanations about its mining decisions to trigger any needed future updates. Another advantage of the NegMiner is that, unlike the NegEx algorithm, it can easily deal with the existence of a UMLS term several times in the same sentence since negated findings in an output sentence are highlighted. Experiments are conducted to show the superiority of the NegMiner in comparison to the NegEx algorithm.
The organization of the paper is as follows: Section II provides details of related research studies in the literature in order to highlight the contributions of the paper. Section III explains the NegEx negation mining algorithm. The details of the proposed NegMiner tool are provided in Section IV. The conducted experiments and the results are presented in Section V. Finally, Section VI presents the conclusion of the paper and directions for future research and improvement of the NegMiner.
-
II. Related Work
Many papers in the literature attempted to tackle the problem of mining negations from electronic narrative medical documents. Algorithms for negation mining can be generally classified into rule-based algorithms, machine leaning algorithms and hybrid algorithms.
One of the earliest rule-based algorithms is the NegExpander algorithm [10-11] that was used to extract absent findings for the ad hoc classification of radiology reports. This syntactic-processing based algorithm processes an input sentence with identified UMLS terms and part-of-speech tags to find conjunctive phrases and then searches for negation phrases inside each conjunctive phrase. Negations inside a given conjunctive phrase are expanded to all UMLS terms inside the phrase. The NegExpander algorithm has been criticized since it does not distinguish between negation phrases preceding and following UMLS terms inside the conjunctive phrases, which may lead to incorrect negations.
One of the most successful rule-based algorithms is the NegEx algorithm [1]. This algorithm utilizes only two regular expressions and a set of negation cues (terms) and termination terms to determine negated UMLS terms. In spite of the simplicity of the NegEx algorithm, its reported mining results are very impressive and in comparison to some classification-based algorithms, it has better agreement with human reviewers [11]. Thus, it has been utilized in several applications and ported to several other natural languages. For example, a negation tagger has been developed based on NegEx to extract information from pathology reports [12]. Meystre and Haug [13] utilized NegEx in a natural language processing system in order to discover any medical conditions in clinical e-documents. NegEx has been ported to the Swedish language [8] and to the French language [9] and its terms lexicon has been extended to multiple languages [3]. Nevertheless, NegEx has been criticized since some negated UMLS terms may be missed and some others may be incorrectly negated. For example, since it originally considered only sentences with at most five tokens between the negation phrases and the UMLS terms, it missed negated UMLS terms in longer sentences [11]. In other words, in spite of the high precision, to be more robust, NegEx needs to be enhanced using lexical and syntactic knowledge to determine the scope of negation phrases [14]. This led to several recent improvements to the NegEx algorithm, but it is still far from being sufficiently robust.
The ConText algorithm [15-16] has been developed as an extension to the NegEx algorithm in order to mine negations in addition to temporal and experiencer statuses from clinical reports. It operates based on a list of conjunctions in order to limit the scopes of the different cues [17]. The frequency of the lexical items utilized by the ConText algorithm was studied [18] and it was observed that about half of those items did not exist in the tested medical documents and thus, it was concluded that trimming the lexical database may have a very limited negative impact. The problem of distinguishing historical findings in clinical reports has been explored further and it was shown that more research need to be made in this respect [19]. In spite of the reported imperfect performance on historical classification [16], PyConText, which is an implementation of a portion of the ConText algorithm in Python has been utilized in many applications. For example, an application called peFinder has been developed based on PyConText for classifying CT pulmonary angiography reports [20]. PyConText has also been used for the automated classification of the history of ancillary cancer of the mesothelioma patients based on free-text clinical reports [21]. The name of PyConText was later changed to PyConTextNLP and it has been ported to the Swedish language [22-23].
Many other rule-based algorithms have been developed. The ENegEx algorithm [24] is an extension to the NegEx algorithm intended to deal with alter-association assertions (associated with someone other than the patient). The SynNeg algorithm [17] uses a syntactic parser to detect the boundaries of sentence units and use them to limit the scope of the negation cues. The NegFinder algorithm [25] was one of the earliest rulebased negation detection algorithms. It utilizes both a lexical scanner with regular expressions and a parser with context-free restricted grammar in order to be able to detect and locate negations [1]. This algorithm has been utilized [26] for finding encounter-based events in clinical electronic medical records and for classifying them. The idea of using syntactic and semantic processing for determining the scope of negation cues has been tackled in many other research studies [27-34].
As mentioned above, machine learning has also been utilized for negation mining. For example, Goldin and Chapman [35] utilized Naïve Bayes and decision tree algorithms to figure out sentences including medical observations negated by the word "not" and sentences that similarly include the word "not" without any negations. Morante et al. [36] utilized k-nearest neighbor classifiers to determine the scope of negation. Rokach et al. [37] designed an algorithm for learning regular expressions using the longest common subsequence algorithm followed by decision tree classification. Morante and Daelemans [38] proposed a machine learning algorithm utilizing k-nearest neighbor classification in addition to Support Vector Machines (SVMs) and conditional random fields to determine the scope of negation. Uzuner et al. [24] proposed the StAC statistical assertion classification algorithm and showed that it outperforms the rule-based ENegEx algorithm. Agarwal and Yu [39] developed the NegScope algorithm that utilizes conditional random fields to detect negations, but it had a lower performance in comparison to NegEx. Fujikawa et al. [40] developed the NegFinder algorithm for determining the scope of negation cues by adding syntactic information to the algorithm of Morante et al. [36, 38] that utilizes k-nearest neighbor classification as discussed above. This algorithm should not be mistaken for the NegFinder algorithm by Mutalik et al. [25].
-
III. The NegEx Algorithm
The original NegEx algorithm was introduced in 2001 [1]. Several modifications have been made to this algorithm since then [4]. In this section, we discuss both versions of the algorithm.
-
A. The Original NegEx Algorithm
The original NegEx algorithm accepts an input sentence with UMLS terms (each replaced by the corresponding UMLS string ID) to determine whether these terms are negated. The algorithm utilizes the following two negation rules (regular expressions):
-
* (2)
In both rules, the asterisk stands for a number of tokens (UMLS terms or merely words) up to five. The algorithm also utilizes two lists of negation phrases. The first list includes pseudo-negation phrases that do not really negate, but indicate double negation (such as not ruled out ), ambiguous negation (such as unremarkable ), or a modified meaning (such as gram-negative ). The second list, on the other hand, includes negation phrases that can be used in the regular expressions above. These negation phrases, in turn, are classified into pre-negation phrases that can be used in rule (1) and post-negation phrases that can be used in rule (2). The original lists include 10 pseudo-negation phrases, 23 pre-negation phrases and only 2 post-negation phrases.
For the algorithm to work, the medical documents are pre-processed so that exactly one sentence appears in each line. Besides, all punctuations (such as commas) are removed. The algorithm proceeds by matching the regular expressions against the sentences in the input document. A possible match of the regular expression (1) is the sentence "The patient denies any kidney pain". On the other hand, a possible match of the regular expression (2) is the sentence "The infection is unlikely ". The asterisk allows matching each regular expression several times against the same sentence. For example, rule (1) is matched twice against the sentence "The patient denies any heart pain or kidney pain" causing the negation of both terms "heart pain" and "kidney pain" by the prenegation phrase denies .
As mentioned before, in spite of the simplicity of the original NegEx algorithm, it has shown impressive mining results, but it still needs some modifications to be more robust. For example, the upper limit of five tokens between the negation phrases and the UMLS terms can lead to missing negated UMLS terms in longer sentences [11]. The negation phrase not is the most problematic. For example, sometimes, it modifies a following non-UMLS term rather than the following UMLS terms. To illustrate, in the sentence "This is not the source of
-
B. The Modified NegEx Algorithm
The NegEx algorithm has been modified [4] by adding a larger number of negation phrases and termination terms that indicate the end of the negation scopes. For example, the termination term but in the sentence "The patient denies any heart pain, but he sweats a lot" indicates the end of the scope of the pre-negation term denies and that it should not extend to the rest of the sentence. This is in addition to the introduction of prepossible-negation terms and post-possible-negation terms. The current lists includes about 16 pseudo-negation terms, 125 pre-negation terms, 21 pre-possible-negation terms, 7 post-negation terms, 14 post-possible-negation terms and 89 termination terms.
With the addition of the termination terms, the asterisk can represent any number of words or UMLS terms in regular expression (1). In this case, the scope of a prenegation term is signaled by the end of the sentence, a termination term or another negation term. On the other hand, the scope of a post-negation term is determined backwards up to five words or UMLS terms. The algorithm starts by searching for pre-negation terms and post-negation terms and then determines the scope of each of these negation terms to identify the negated UMLS terms. In spite of all these modifications, the NegEx algorithm is still not fully robust since many sentences are more complex to be correctly processed by just a couple of rules and a set of relatively few negation and termination terms.
-
IV. The NegMiner Tool
As shown in Figure 1, the NegMiner tool is formed of three modules: the knowledge base, the update module and the mining module. The knowledge base includes the negation cues, the termination terms and the mining rules. The update module is responsible for facilitating the process of updating the knowledge base and the mining module is responsible for the negation mining process. We explain the details of each of those modules in the following sub-sections.
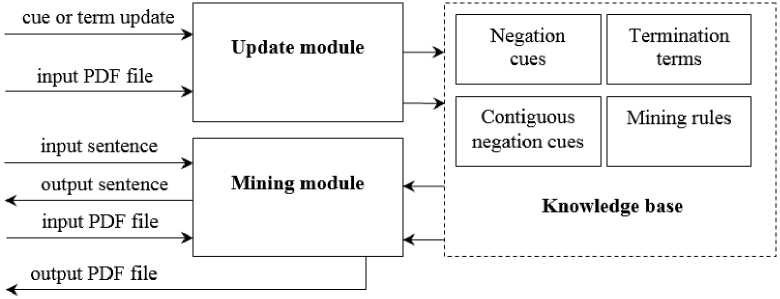
Fig.1. Block diagram of the NegMiner tool.
-
A. The Knowledge Base
As mentioned above, the knowledge base of the NegMiner is responsible for storing the negation terms, the termination terms and the mining rules. The NegMiner utilizes the same negation terms and termination terms as the NegEx algorithm with few modifications such as adding the term deny to the list of pre-negation terms. We also added a set of contiguous negation terms that are not considered unless contiguous to UMLS terms. These are, in turn, classified into pre-contiguous negation terms that precede the UMLS terms and post-contiguous negation terms that follow the UMLS terms. Examples of pre-contiguous negation terms are absent and negative while examples of post-contiguous negation terms are absence and also negative . These contiguous negation terms are intended to detect some common negations that are, unfortunately not considered by the NegEx algorithm.
The mining rules utilized by the NegMiner are different from those of the NegEx algorithm to be able to deal with contiguous and multiple negations. Unlike the negation rules of the NegEx algorithm, the mining rules of the NegMiner do not negate the corresponding UMLS terms, but change their statuses. Each UMLS term is initially existent. Whenever one of the rules is matched against a sentence fragment including a UMLS term, the status of the term is changed from existent to negated or vice versa. The NegMiner mining rules are as follows:
(
*
In these rules, the asterisk stands for any number of words, UMLS terms or contiguous negation terms. On the other hand, the hash stands for up to five words excluding conjunctions such as and. The power, n is any positive integer or 0. When n = 0, rules (5) and (6) are equivalent to rules (1) and (2) of the NegEx algorithm. A value of n > 0 indicates the occurrence of some words preceded by a pre-negation term n times.
-
B. The Mining Module
The mining module is responsible for mining negations using the knowledge base. Like the NegEx algorithm, the NegMiner accepts an input sentence with identified UMLS terms and processes it searching for negated UMLS terms. To process an input sentence, unlike the NegEx algorithm, the NegMiner does not start with the negation cues, but starts with the UMLS terms instead. When a UMLS term is encountered in an input sentence, the sentence is first matched against rules (3) and then (4). If a pre-contiguous or a post-contiguous negation term is encountered, the status of the UMLS term is changed from existent to negated. Next, each of rules (5) and (6) is matched as many times as n+1. Whenever any of these rules matches, the status of the UMLS term is changed accordingly. In other words, assuming no contiguous negation terms, a UMLS term is negated only in case of an even value of n.
The mining module can process individual sentences. But, to simplify the mining process, it accepts an input PDF file with multiple sentences, one per line. It processes the file and outputs a PDF file with the mining results. Generally, the mining module outputs processed sentences with highlighted negated UMLS terms. An explanation accompanies each UMLS term (whether negated or not) explaining the mining decision. In case of an output PDF file, explanations are added as comments to the UMLS terms. This has two advantages. First, explanations highlight shortcomings in the mining rules, which can trigger future enhancements. Second, this helps the NegMiner in dealing with one of the prominent problems of the NegEx algorithm, which is its inability to deal with the existence of the same UMLS term several times in one sentence. If such a UMLS term is negated once, it is reported by the NegEx algorithm as being generally negated even if its other occurrences are not negated. The NegMiner, on the other hand, deals with each occurrence of a given UMLS term separately and highlights only the negated ones.
-
C. The Update Module
The function of the update module is to facilitate the process of updating the knowledge base. The NegMiner is a user-friendly tool that allows the user to browse the negation and termination terms and update them as required. It also allows deleting any of them and adding new ones. The update module also accepts input PDF files including sentences with highlighted words and comments added to indicate the class of each. This information can be extracted from the file for the automatic update of the knowledge base without having to do this manually one by one.
-
V. Experiments and Results
In order to evaluate the NegMiner algorithm, we prepared 500 sentences for the experiments. We made sure that all the negation phrases and the UMLS terms that appeared in the sentences were already identified. We evaluated both the simulated NegEx algorithm and the NegMiner algorithm using the same set of sentences and compared them in terms of Positive Predictive Value (PPV) and Negative Predictive Value (NPV) in addition to Sensitivity and Specificity that are defined in equations (7) through (10) [41]:
Positive Predictive Value = Σ TP / ( Σ TP + Σ FP) (7)
Negative Predictive Value = Σ TN / ( Σ TN + Σ FN) (8)
Sensitivity = Σ TP / ( Σ TP + Σ FN) (9)
Specificity = Σ TN / ( Σ TN + Σ FP) (10)
The True Positives (TP) are terms negated by the raters and by the system, the True Negatives (TN) are terms that are not negated by both the raters and the system, the False Positives (FP) are terms negated by the system and not by the raters and finally, the False Negatives (FN) are terms negated by the raters and not by the system [1].
The outcomes of the conducted experiments are provided in Table 1. From these results, it is obvious that the NegMiner algorithm has superior performance in comparison with the simulated NegEx algorithm. This is due to the design of the NegMiner and its superior capabilities. To illustrate, examples are provided in the following sub-sections to demonstrate the strengths and weaknesses of the NegMiner in comparison to the simulated NegEx algorithm.
Table 1. Evaluation of the mining results of the NegMiner algorithm in comparison to the simulated NegEx algorithm
|
Evaluation Criteria |
NegEx |
NegMiner |
|
Sensitivity |
93% |
95% |
|
Specificity |
90% |
95% |
|
Positive Predictive Value |
86% |
93% |
|
Negative Predictive Value |
95% |
96% |
-
A. Multiple Occurrences of a UMLS Term
As mentioned before, the NegEx algorithm is unable to deal with sentences in which there is more than one occurrence of a given UMLS term possibly resulting in false positives in case of at least a single negation. This problem is not existent in the NegMiner that treats UMLS terms separately and highlights only negated ones. To illustrate, consider the sentence [1] "The patient was subject to precautions of
-
B. Single Negation
The sentence "The patient
denies
having
-
C. Double Pre-negation
The sentence "The patient did
not deny
having
The sentence “We could
not
confirm the
absence of
-
D. Pre-negation and Post-negation
The sentence "We
cannot
say that having
-
E. Double Post-negation
The sentence "Being
-
F. Contiguous Negation and Several UMLS Terms
The sentence "We
cannot deny
that being
In the sentence "The patient has
no
On the other hand, in the sentence "The patient has
no
-
G. Sentences Requiring Semantic Information
In spite of the ability of the NegMiner to provide improved performance in comparison to the NegEx algorithm, more effort still needs to be done to handle more complex sentences. For example, the two sentences “We did
not
treat
The sentence “The heart EKG showed
no
-
VI. Conclusion
This paper proposes the novel NegMiner tool to address some of the shortcomings of the popular and widely-adopted NegEx algorithm. The NegMiner exploits some basic syntactic and semantic information to deal with more negations in comparison to the NegEx algorithm. Thus, it considers pre- and post-contiguous negation cues. Besides, we have updated the rules of the NegEx algorithm to be able to deal with multiple negations. The NegMiner is a user-friendly tool that facilitates the task of knowledge base update. It also facilitates the mining process since it accepts individual sentences or an input PDF file including a set of sentences. Each UMLS term in an output sentence is accompanied by explanation of the mining decision to help highlight any shortcomings that would trigger future updates. This capability also helps in addressing one of the prominent problems of the NegEx algorithm, which is its inability to deal with the existence of a UMLS term several times in a single sentence.
Experimental results have shown a superior performance of the NegMiner algorithm in comparison to the simulated NegEx algorithm. It was clear that the NegMiner would have performed better if we could widen the scope of the contiguous negation terms and constraint the negation terms that could be applied together. This will be considered as a future work. Finally, some additional syntactic and semantic information might be needed to deal with more complex sentence structures.
It should be noted that the NegEx algorithm has been simulated based on the limited information provided [4], since we could not find a recent formal publication describing the modified algorithm. It should be also noted that the results provided in this paper are based on the random sentence set utilized. An important point of future research would be a benchmark classification of different forms of sentences and negations that appear in medical documents so that each algorithm would report its capabilities based on the classes of possible sentences and negations rather than on a random set. We are currently working on this point of research. Finally, we intend to port the NegMiner to other natural languages.
Список литературы NegMiner: An Automated Tool for Mining Negations from Electronic Narrative Medical Documents
- F. Chapman, W. Bridewell, P. Hanbury, G. Cooper, and B. Buchanan, "A simple algorithm for identifying negated findings and diseases in discharge summaries," Journal of Biomedical Informatics, vol. 34, pp. 301-310, 2001.
- Unified Medical Language System (UMLS), https://www.nlm.nih.gov/research/umls/, [Online; accessed: 2016-06-01].
- W. Chapman, D. Hilert, S. Velupillai, M. Kvist, M. Skeppstedt, B. Chapman, M. Conway, M. Tharp, D. Mowery, and L. Deleger, "Extending the NegEx lexicon for multiple languages," Studies in Health Technology and Informatics, vol. 192, pp. 677-681, 2013.
- Negex, https://code.google.com/p/negex/, [Online; accessed: 2016-06-01].
- R. Morante, "Descriptive analysis of negation cues in biomedical texts," Proceedings of the 7th Conference on International Language Resources and Evaluation, Valletta, Malta, pp. 1429-1436, 2010.
- Y. Huang and H. Lowe, "A grammar-based classification of negations in clinical radiology reports," Proceedings of AMIA Symposium, Washington, DC, USA, p. 988, 2005.
- V. Vincze, G. Szarvas, R. Farkas, G. Mora, and J. Csirik, "The BioScope Corpus: Biomedical texts annotated for uncertainty, negation and their scopes," BMC Bioinformatics, vol. 9, no. 11, 2008.
- M. Skeppstedt, "Negation detection in Swedish clinical text: An adaption of NegEx to Swedish," Journal of Biomedical Semantics, vol. 2, no. 3, 2011.
- L. Deleger and C. Grouin, "Detecting negation of medical problems in French clinical notes," Proceedings of the 2nd ACM SIGHIT International Health Informatics Symposium, Miami, Florida, pp. 697-702, 2012.
- D. Aronow, F. Fangfang, and W. Croft, "Ad hoc classification of radiology reports," Journal of the American Medical Informatics Association, vol. 6, no. 5, pp. 393-411, 1999.
- S. Goryachev, M. Sordo, Q. Zeng, and L. Ngo, "Implementation and evaluation of four different methods of negation detection," Technical Report, Harvard Medical School, Boston, USA, 2006.
- K. Mitchell, M. Becich, J. Berman, W. Chapman, J. Gilbertson, D. Gupta, J. Harrison, E. Legowski, and R. Crowley, "Implementation and evaluation of a negation tagger in a pipeline-based system for information extraction from pathology reports," Proceedings of MEDINFO, pp. 663-667, 2004.
- S. Meystre and P. Haug, "Natural language processing to extract medical problems from electronic clinical documents: Performance evaluation," Journal of Biomedical Informatics, vol. 39, pp. 589-599, 2006.
- W. Chapman, W. Bridewell, P. Hanbury, G. Cooper, and B. Buchanan, "Evaluation of negation phrases in narrative clinical reports," Proceedings of the AMIA Symposium, Washington, DC, USA, pp. 105-109, 2001.
- W. Chapman, D. Chu, and J. Dowling, "ConText: An algorithm for identifying contextual features from clinical text," Proceedings of the Workshop on BioNLP 2007: Biological, Translational and Clinical Language Processing, Prague, Czech Republic, pp. 81-88, 2007.
- H. Harkema, J. Dowling, T. Thornblade, and W. Chapman, "ConText: An algorithm for determining negation, experiencer, and temporal status from clinical reports," Journal of Biomedical Informatics, vol. 42, pp. 839-851, 2009.
- H. Tanushi, H. Dalianis, M. Duneld, M. Kvist, M. Skeppstedt, and S. Velupillai, "Negation scope delimitation in clinical text using three approaches: NegEx, PyConTextNLP and SynNeg," Proceedings of the 19th Nordic Conference of Computational Linguistics, Oslo, Norway, pp. 387-474, 2013.
- B. Chapman, W. Wei, and W. Chapman, "The frequency of ConText lexical items in diverse medical texts," Proceedings of the IEEE 2nd Conference on Healthcare Informatics, Imaging and Systems Biology, La Jolla, California, USA, p. 135, 2012.
- D. Mowery, H. Harkema, J. Dowling, J. Jonathan, L. Lustgarten, and W. Chapman, "Distinguishing historical from current problems in clinical reports - Which textual features help?" Proceedings of the Workshop on BioNLP, Boulder, Colorado, USA, pp. 10-18, 2009.
- B. Chapman, S. Lee, H. Kang, and W. Chapman, "Document-level classification of CT pulmonary angiography reports based on an extension of the ConText algorithm," Journal of Biomedical Informatics, vol. 44, pp. 728-737, 2011.
- R. Wilson, W. Chapman, S. DeFries, M. Becich, and B. Chapman, "Automated ancillary cancer history classification for mesothelioma patients from free-text clinical reports," Journal of Pathology Informatics, vol. 1, no. 24, 2010.
- S. Velupillai, M. Skeppstedt, M. Kvist, D. Mowery, B. Chapman, H. Dalianis, and W. Chapman, "Porting a rule-based assertion classier for clinical text from English to Swedish," Proceedings of the 4th International Louhi Workshop on Health Document Text Mining and Information Analysis, Sydney, Australia, 2013.
- S. Velupillai, M. Skeppstedt, M. Kvist, D. Mowery, B. Chapman, H. Dalianis, and W. Chapman, "Cue-based assertion classification for Swedish clinical text - Developing a lexicon for pyConTextSwe," Artificial Intelligence in Medicine, vol. 61, pp. 137-144, 2014.
- Ö. Uzuner, X. Zhang, and T. Sibanda, "Machine learning and rule-based approaches to assertion classification," Journal of the American Medical Informatics Association, vol. 16, no. 1, pp. 109-115, 2009.
- P. Mutalik, A. Deshpande, and P. Nadkarni, "Use of general-purpose negation detection to augment concept indexing of medical documents: A quantitative study using the UMLS," Journal of the American Medical Informatics Association, vol. 8, no. 6, pp. 598-609, 2001.
- B. Hazlehurst, H. Frost, D. Sittig, and V. Stevens, "MediClass: A system for detecting and classifying encounter-based clinical events in any electronic medical record," Journal of the American Medical Informatics Association, vol. 12, no. 5, pp. 517-529, 2005.
- S. Boytcheva, A. Strupchanska, E. Paskaleva, and D. Tcharaktchiev, "Some aspects of negation processing in electronic health records," Proceedings of the International Workshop on Language and Speech Infrastructure for Information Access in the Balkan Countries, Borovets, Bulgaria, 2005.
- H. Tolentino, M. Matters, W. Walop, B. Law, W. Tong, F. Liu, P. Fontelo, K. Kohl, and D. Payne, "Concept negation in free text components of vaccine safety reports," Proceedings of AMIA Symposium, Washington, DC, USA, p. 1122, 2006.
- Y. Huang and H. Lowe, "A novel hybrid approach to automated negation detection in clinical radiology reports," Journal of the American Medical Informatics Association, vol. 14, no. 3, pp. 304-311, 2007.
- S. Gindl, K. Kaiser, and S. Miksch, "Syntactical negation detection in clinical practice guidelines," Studies in Health Technology and Informatics, vol. 136, pp. 187-192, 2008.
- Q. Zhu, J. Li, H. Wang, and G. Zhou, "A unified framework for scope learning via simplified shallow semantic parsing," Proceedings of the 2010 Conference on Empirical Methods in Natural Language Processing, MIT, Massachusetts, USA, pp. 714-724, 2010.
- M. Ballesteros, V. Francisco, A. Diaz, J. Herrera, and P. Gervas, "Inferring the scope of negation in biomedical documents," Proceedings of the 13th International Conference on Intelligent Text Processing and Computational Linguistics, New Delhi, India, 2012.
- E. Velldal, L. Øvrelid, J. Read, and S. Oepen, "Speculation and negation: Rules, rankers, and the role of syntax," Computational Linguistics, vol. 38, no. 2, pp. 369-410, 2012.
- Z. Jia, H. Li, M. Ju, Y. Zhang, Z. Huang, C. Ge, and H. Duan, "A finite-state automata based negation detection algorithm for Chinese clinical documents," Proceedings of International Conference on Progress in Informatics and Computing, Shanghai, China, pp. 128-132, 2014.
- I. Goldin and W. Chapman, "Learning to detect negation with ‘not’ in medical texts," Proceedings of the 26th Annual International ACM SIGIR Conference on Research and Development on Information Retrieval, Toronto, Canada, 2003.
- R. Morante, A. Liekens, and W. Daelemans, "Learning the scope of negation in biomedical texts," Proceedings of the 2008 Conference on Empirical Methods in Natural Language Processing, Honolulu, Hawaii, USA, pp. 715-724, 2008.
- L. Rokach, R. Romano, and O. Maimon, "Negation recognition in medical narrative reports," Information Retrieval, vol. 11, pp. 499-538, 2008.
- R. Morante and W. Daelemans, "A Metalearning approach to processing the scope of negation," Proceedings of the 13th Conference on Computational Natural Language Learning, Boulder, Colorado, USA, pp. 21-29, 2009.
- S. Agarwal and H. Yu, "Biomedical negation scope detection with conditional random fields," Journal of the American Medical Informatics Association, vol. 17, pp. 696-701, 2010.
- K. Fujikawa, K. Seki, and K. Uehara, "NegFinder: A web service for identifying negation signals and their scopes," Technical Report, IPSJ SIG, 2013.
- S. Gindl, "Negation detection in automated medical applications: A Survey," Technical Report TR-2006-1, Institute of Software Technology & Interactive Systems, Vienna University of Technology, 2006.
- I. Khan and M. Haleem, "Managing Lexical Ambiguity in the Generation of Referring Expressions," International Journal of Intelligent Systems and Applications, no. 8, pp. 33-39, 2013.


