Практическое применение массового параллельного репортерного анализа в биотехнологии и медицине
Автор: Романов Станислав Евгеньевич, Лактионов Петр Павлович
Журнал: Клиническая практика @clinpractice
Рубрика: Научные обзоры
Статья в выпуске: 4 т.13, 2022 года.
Бесплатный доступ
Развитие и жизнедеятельность организма опирается на тканеспецифичные генные программы. Ключевую роль в регуляции таких программ играют регуляторные элементы генома, а нарушения в их работе могут приводить к развитию различных патологий, включая пороки развития, онкологические и аутоиммунные заболевания. Развитие высокопроизводительных геномных исследований привело к появлению методов массового параллельного репортерного анализа (МПРА), которые позволяют проводить функциональную проверку и идентификацию регуляторных элементов в масштабе генома. Изначально МПРА применялся в качестве инструмента для исследования фундаментальных аспектов эпигенетики, однако этот подход также имеет огромный потенциал для клинической и практической биотехнологии. На текущий момент МПРА используют для валидации клинически значимых мутаций, идентификации тканеспецифичных регуляторных элементов, поиска наиболее перспективных для интеграции трансгена локусов. МПРА является также незаменимым инструментом для создания высокоэффективных экспрессионных систем, спектр применения которых распространяется от подходов для наработки белков и конструирования суперпродуцентов терапевтических антител нового поколения до генной терапии. В настоящем обзоре предложены к рассмотрению основные принципы и области практического применения методов высокопроизводительного репортерного анализа.
Массовый параллельный репортерный анализ, мпра, экспрессионные системы, биотехнология, клиническая биоинженерия
Короткий адрес: https://sciup.org/143179874
IDR: 143179874 | DOI: 10.17816/clinpract115063
Текст обзорной статьи Практическое применение массового параллельного репортерного анализа в биотехнологии и медицине
В основе развития и жизнедеятельности организма лежат тканеспецифичные генные программы, которые контролируются многоуровневыми взаимодействиями между белковыми комплексами, влияющими на транскрипцию, и специальными последовательностями ДНК — цис-регуляторными элементами (ЦРЭ) [1, 2]. Нарушения такой коммуникации могут приводить к различной патологии, включая онкологические заболевания, пороки развития и аутоиммунные заболевания [3–7].
Современные методы геномного анализа существенно расширили представления о генетических и эпигенетических свойствах регуляторных элементов, что позволяет идентифицировать потенциальные ЦРЭ в различных типах клеток [8]. Таким образом были выявлены десятки тысяч геномных локусов модельных организмов и человека, потенциально вовлеченных в регуляцию экспрессии генов [9]. Подобные исследования значительно расширяют представления о механизмах тонкой регуляции экспрессии генов как в норме, так и при патологии [4, 10].
Помимо фундаментальных аспектов, идентификация и описание функций отдельных регуляторных элементов генома позволяют применять их в создании экспрессионных конструкций для биотехнологии, например, при наработке белковых препаратов или в подходах генной терапии [11, 12]. Одним из ключевых требований к экспрессион-ным генетическим конструкциям в подобных применениях является отсутствие генотоксичности,
Лицензия CC BY-NC-ND 4 / The article can be used under the CC BY-NC-ND 4 license
PRACTICAL APPLICATION OF MASSIVELY PARALLEL REPORTERASSAY IN BIOTECHNOLOGY AND MEDICINE
S.E. Romanov1, 2, P.P. Laktionov1, 2
-
1 Novosibirsk State University, Novosibirsk, Russian Federation
-
2 Institute of Molecular and Cellular Biology, Siberian Branch of the Russian Academy of Sciences, Novosibirsk, Russian Federation
The development and viability of an organism relies on tissue-specific gene programs. The genome regulatory elements play a key role in the regulation of such programs, whereas its disfunction can lead to the establishment of various pathologies, including cancer, congenital disorders, and autoimmune diseases. The development of high-throughput approaches in genomics has led to the emergence of massively parallel reporter assays (MPRA), which enable genome-wide screening and functional verification of regulatory elements. Although MPRA was originally used for investigation of fundamental aspects of epigenetics, it also has a great potential for clinical and practical biotechnology. Currently, MPRA is used for validation of clinically significant mutations, identification of tissue-specific regulatory elements, identification of the favorable loci for transgene integration, as well as represents an essential tool for creating highly efficient expression systems, with a wide range of applications from protein production and design of novel therapeutic antibody super-producers to gene therapy. In this review, the basic principles and areas of practical application of high-throughput reporter assays will be discussed. Keywords: massively parallel reporter assay; MPRA; expression systems; biotechnology; clinical bioengineering.
doi:
Submitted 05.11.2022 Revised 13.12.2022 Published 30.12.2022
а также обеспечение стабильной и контролируемой экспрессии, что невозможно без рационального дизайна, включающего подбор адекватных регуляторных элементов [13, 14]. Вместе с тем известные регуляторные последовательности зачастую не лишены практических недостатков, а выбор иных регуляторных элементов требует их функционального тестирования.
Для осуществления такого тестирования в масштабе генома были разработаны методы массового параллельного репортерного анализа (МПРА), которые позволяют в ходе одного эксперимента проанализировать тысячи вариантов потенциальных регуляторных последовательностей [15]. Менее чем за десять лет данная методология способствовала значительному прогрессу как в исследовании фундаментальных принципов функционирования регуляторных элементов генома, так и в их практическом применении в подходах медицинской генетики и генной инженерии.
В обзоре рассматриваются основные стратегии МПРА для идентификации различных регуляторных элементов в геноме и его практическое применение.
АНАЛИЗ ЦИС-РЕГУЛЯТОРНЫХЭЛЕМЕНТОВ ГЕНОМА
Основные классы регуляторныхэлементов генома
Регуляторные элементы генома представляют собой короткие участки генома, свободные от нуклеосом и доступные для связывания регуляторных белков [16]. В зависимости от регуляторного эффекта выделяют несколько типов ЦРЭ [17–20].
Промоторы представляют собой проксимальные ЦРЭ, расположенные вблизи сайта инициации транскрипции генов [17]. На промоторах происходит сборка транскрипционного комплекса, состоящего из РНК-полимеразы, главных факторов транскрипции и иных регуляторных белков, в том числе опосредующих взаимодействие с энхансерами [17, 21].
Энхансеры представлены короткими фрагментами ДНК, служащими сайтами посадки транскрипционных факторов и кофакторов. В составе такого белкового комплекса энхансеры способны физически контактировать с промотором гена, что приводит к его активации [18]. Энхансеры явля-


ются дистальными ЦРЭ и способны активировать экспрессию целевого гена, находясь на удалении вплоть до миллиона пар нуклеотидов, вне зависимости от пространственной ориентации с промотором [22, 23]. В среднем в геноме позвоночных энхансеры удалены на 20–50 т.п.н. (тысяч пар нуклеотидов) от целевых генов [18].
Взаимодействия энхансеров с промоторами лежат в основе комплексной тканеспецифичной регуляции генов, в которой отдельный ген может быть подвержен регуляции несколькими энхансерами, иметь общие энхансеры с другими генами и подвергаться регуляции специфичными энхансерами в разных типах клеток [18, 24]. При развитии патологий наблюдается также регуляция генов специфичными энхансерами. Так, например, энхансер протоонкогена Myc располагается в районе сайта терминации транскрипции при раке поджелудочной железы, при раке прямой кишки обнаруживается со стороны 5’-области гена, а в случае T-клеточной острого лимфобластного лейкоза — ниже 3’-области гена [25].
Инсуляторы. Дистальное взаимодействие энхансеров с промоторами целевых генов обеспечивается за счет формирования петель хроматина, в том числе под действием моторных белков, таких как РНК-полимераза II и когезин, ключевую же роль в их организации играют инсуляторы — отдельный класс ЦРЭ. Исходно инсуляторы были охарактеризованы как последовательности ДНК, способные блокировать взаимодействие между промоторами и энхансерами, а также формировать барьеры на границе гетерохроматина и эухроматина [26, 27]. Изучение пространственной структуры ядра позволило установить, что функциональное влияние инсуляторов во многом определяется их участием в регуляции внутри- и межхромосомных контактов [20]. У млекопитающих инсуляторы служат сайтами связывания CCCTC-связывающего белка CTCF, который способен к гомодимеризации, что приводит к формированию петель хроматина [28, 29]. В результате подобных взаимодействий формируются обособленные друг от друга участки генома протяженностью порядка сотен тысяч пар нуклеотидов — топологически ассоциированные домены [30]. Такие структуры характеризуются более высокой частотой внутренних промотор-энхансерных взаимодействий, а нарушение их структуры наблюдается при различных патологиях, таких как онкологические заболевания и болезни, ассоциированные с экспансией нуклеотидных повторов [30–34].
Сайленсеры являются дистальными регуляторными элементами, функция которых заключается в подавлении экспрессии генов [19]. Такие элементы являются сайтами посадки белков-репрессоров транскрипции, которые реализуют функциональное влияние посредством установления репрессивных хроматиновых меток и конкуренции с активирующими белками [35–40]. Зачастую сайленсеры располагаются рядом с энхансерами или даже пересекаются с ними [41]. Действие таких бифункцио- нальных регуляторных элементов на транскрипцию зависит от эпигенетического контекста и обеспечивает тонкую настройку активности генов посредством конкуренции между репрессорными и актива-торными факторами [41].
Подходы к идентификации регуляторных элементов in silico
Современные методы геномного анализа значительно расширили представления о характерных генетических, эпигенетических и структурных особенностях разных классов регуляторных элементов, что в свою очередь позволяет идентифицировать потенциальные ЦРЭ. Для предсказания потенциальной регуляторной роли геномной последовательности необходимо оценить доступность ДНК для белковых факторов, проанализировать представленность характерных эпигенетических меток и белков, изучить пространственную структуру хроматина исследуемой области и оценить ее влияние на экспрессию генов. Основные эпигенетические детерминанты, ассоциированные с ЦРЭ, и методы их идентификации перечислены в табл. 1 [30, 48–76].
С использованием указанных в табл. 1 подходов в геномах модельных организмов и человека обнаружены тысячи регуляторных элементов, каталоги которых можно найти в различных геномных базах данных [42–47]. Вместе с тем, учитывая технические особенности методов идентификации потенциальных ЦРЭ, вариабельность функций эпигенетических детерминант и участие множества компонентов в реализации функций элементов генома in vivo , валидация регуляторной роли ЦРЭ требует функциональной проверки.
Массовый параллельный репортерный анализ для исследования цис-регуляторных элементов
Классической функциональной проверкой регуляторных элементов генома является репортерный анализ, при котором оценивается уровень экспрес-
Таблица 1 / Table 1
Эпигенетические детерминанты, ассоциированные с цис-регуляторными элементами, и методы их идентификации
Epigenetic determinants associated with cis-regulatory elements and methods for their identification
|
Эпигенетические детерминанты |
Особенности ЦРЭ |
Методы исследования |
|
Модификации гистонов, транскрипционные факторы |
Промоторы: H3K4me3 [48]. Активные энхансеры: H3K4me1, H3K27ac, P300 [49, 50]. Неактивные энханcеры: H3K4me1, H3K27me3 [51]. Суперэнхансеры: H3K27ac, Mediator, Oct4, Sox2, Nanog, Klf4 [52]. Сайленсеры: H3K27me3, H3K79me2, H4K20me1, EZH2 [53, 54]. Инсуляторы: CTCF, Rad21, YY [30, 55–57] |
ChIP-seq [58] ChIP-exo [59] |
|
Доступность хроматина |
Активные энхансеры [60], сайленсеры [61] и инсуляторы [62, 63] располагаются в областях свободного от нуклеосом открытого хроматина. Промоторы активных генов совпадают с областями открытого хроматина и демонстрируют характерный паттерн распределения нуклеосом [64] |
DNase-seq [65] MNase-seq [66] ATAC-seq [67] FAIRE-seq [68] |
|
Пространственная организация хромосом |
Энхансеры [69] и сайленсеры [54, 61, 70] физически взаимодействуют с промоторами при помощи петлевых контактов. Инсуляторы формируют петлевые домены, ограничивающие область действия энхансеров [71, 72] |
HiC [73] ChIA-PET [74] HiChIP [75] PLAC-ChIP [76] |
сии маркерного гена, находящегося под контролем исследуемого ЦРЭ в составе генетической конструкции [77]. Однако анализ даже нескольких сотен потенциальных регуляторных элементов является крайне ресурсоемким, так как подразумевает проведение соответствующего количества индивидуальных экспериментов [78].
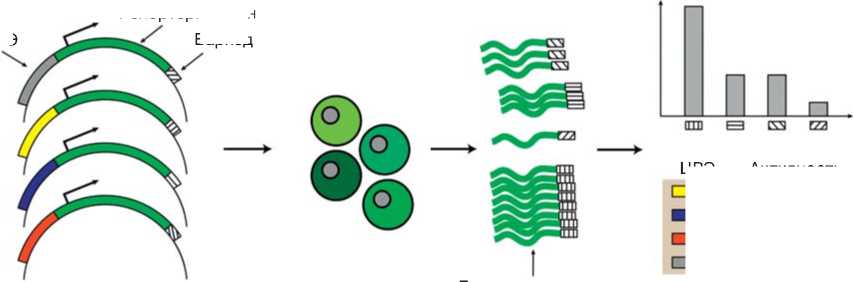
Развитие методов высокопроизводительного секвенирования позволило преодолеть это ограничение и проводить одновременный анализ активности множества регуляторных элементов. Ключевым отличием методов массового параллельного репортерного анализа (МПРА) в сравнении с классическими подходами является внедрение в транскрибируемую 3’-концевую область репортерного гена коротких вырожденных последовательностей — ДНК-баркодов [79] (рис. 1, а ). Для проведения МПРА проводится трансгенез с использованием пула из тысяч репортерных конструкций, каждая из которых содержит потенциальный регуляторный элемент и репортерный ген с уникальным ДНК-баркодом. Анализ представленности ДНК-баркодов в транскриптах репортерного гена методом высокопроизводительного секвенирования позволяет сделать вывод о регуляторной активности соответствующего ему ЦРЭ [80–82].
На текущий момент разработано множество количественных и качественных вариаций методов МПРА, отличающихся способами подготовки анализируемых ДНК-фрагментов и способами трансгенеза. Более подробную информацию о вариаци- ях метода МПРА можно найти в опубликованных ранее работах [15, 83].
Первые вариации систем для МПРА-анализа использовали минимальный промотор, который, в отсутствие активирующего влияния ЦРЭ, поддерживает уровень экспрессии репортерного гена на крайне низком уровне. По этой причине в первую очередь МПРА применялся для de novo идентификации энхансеров или изучения влияния мутаций на известные активирующие ЦРЭ [80, 84–86]. Аналогичные подходы были использованы для исследования промоторов, однако для этого в состав репортерных конструкций помещали репортерный ген без корового промотора [87]. Использование же активного промотора в составе репортерной конструкции позволило исследовать функцию сай-ленсеров путем оценки относительного снижения уровня представленности ДНК-баркодов по сравнению с контрольными конструкциями [61, 88].
Элегантный подход для идентификации сайлен-серов в масштабе генома был использован в работе [54] (рис. 1, б ). Для этого фрагменты геномной ДНК были клонированы в состав генетических конструкций перед геном, кодирующим химерный белок FKBP-Casp9, под управлением активного промотора EF1 α . Химерный белок состоит из модифицированной каспазы-9 и FK506-связывающе-го белка и под воздействием индуктора AP20187 способен димеризоваться и индуцировать апоптоз [89, 90]. Если экспрессионная кассета попадает под воздействие сайленсера, то экспрессия гена

Клонирование пула баркодированных конструкций
Трансформация целевых клеток
Выделение мРНК
Секвенирование баркодов + оценка активности ЦРЭ
Репортерный ген Баркод
Баркодированные транскрипты репортера
ЦРЭ Активность
ЦРЭ
a
Потенциальные сайленсеры
Клонирование в конструкции
Трансформация целевых клеток
Индукция апоптоза
Выделение ДНК + Прямое секвенирование ЦРЭ
б
AP20187
Конструкция для экспрессии суицидного белка FKBP-Casp9
МПРА для исследования посттранскрипционной регуляции генов
БК полиА]-
МПРА для исследования сплайсинга
Интрон
mCherry
1 GFP ZZb
Зрелый транскрипт
в

J mCherryJБКш::п GFP mCherry GFP mCherryjБК m GFPzj^ О + О
mCherry

Рис. 1. Стратегии массового параллельного репортерного анализа (МПРА): а — принцип МПРА (сначала набор выбранных ЦРЭ клонируется в пул репортерных конструкций, каждая из которых несет уникальный баркод; после трансформации целевых клеток из них выделяется РНК, и представленность баркодов в транскриптах репортера используется как мера активности ЦРЭ); б — схема метода ReSE для высокопроизводительного поиска сайлен-серов (см. пояснения в тексте); в — для исследования посттранскрипционной регуляции генов в баркодированную (БК) репортерную конструкцию помещаются последовательности 5’- или 3’- нетранслируемой области (НТО), либо терминатор транскрипции с сигналом полиаденилирования; для изучения сплайсинга в баркодированную конструкцию помещаются варианты интронов, фланкированных разными сплайс-сайтами; нарушение сплайсинга сопровождается сдвигом содержания репортерных белков в клетках (GFP и mCherry); далее клетки можно разбить на группы по уровню активности репортерных белков и проанализировать обогащение баркодов в каждой группе. Fig. 1. Strategies for massively parallel reporter assay: а — Principle of MPRA. Initially, a set of selected CREs is cloned into a pool of reporter constructs, each carrying a unique barcode. After transformation, RNA is extracted from the cells, and barcodes in the reporter transcripts is used as a measure of CRE activity; б — Schematic of the ReSE method for high-throughput detection of silencers (see explanations in the text); в — To study the post-transcriptional regulation of genes, the 5' or 3'- untranslated region (UTR) sequences or a transcription terminator with a polyadenylation signal are placed in the barcoded (BC) reporter construct. To investigate splicing, variants of introns flanked by different splice sites are placed in the barcoded construct. Disruption of splicing is accompanied by a shift in the content of the reporter proteins in the cells (GFP and mCherry). Next, cells can be categorized into groups based on the level of reporter protein activity, and barcode enrichment in each group can be analyzed.
химерного белка подавляется, а клетки, содержащие такую конструкцию, сохраняют жизнеспособность при химической индукции апоптоза. Таким образом, в пуле трансфицированных клеток выживают только несущие генетические конструкции, содержащие сайленсеры. Последующее таргетное высокопроизводительное секвенирование позволяет установить последовательности сайленсеров. Представленный метод является одним из первых подходов для систематической высокопроизводительной идентификации сайленсеров [54].
Возможности массового параллельного репор-терного анализа не ограничиваются поиском промоторов, энхансеров и сайленсеров. Существуют системы МПРА, позволяющие анализировать регуляторный эффект комбинаций нескольких ЦРЭ [91, 92]. В частности, недавно это позволило исследовать регуляторную функцию потенциальных ин-суляторов в геномном контексте [93]. Кроме того, к настоящему времени предложено множество различных модификаций МПРА для исследования последовательностей, вовлеченных в сплайсинг мРНК [94–96], регуляцию трансляции белков и стабильности мРНК [97–99] и посттранскрипционные модификации РНК [100, 101] (рис. 1, в ).
ОБЛАСТИ ПРАКТИЧЕСКОГО ПРИМЕНЕНИЯ МАССОВОГО ПАРАЛЛЕЛЬНОГО РЕПОРТЕРНОГО АНАЛИЗА
Применение массового параллельного репортерного анализа для оптимизации экспрессионных конструкций
Обеспечение заданного уровня и стабильности экспрессии ДНК-трансгенов является одной из принципиальных технических задач в различных биотехнологических подходах, таких как генная терапия или продукция терапевтических белков. Однако доступные в настоящий момент системы не лишены недостатков. Например, часто используемые вирусные промоторы CMV и SV40 имеют высокую активность в S-фазе, которая снижается после индуцированного покоя клеток [102–106]. В связи с этим индукция дифференцировки, которая в ряде случаев приводит к остановке клеточного цикла, может приводить к снижению экспрессии трансгенов, что является проблемой для генной терапии [107, 108]. Для промотора CMV также характерна инактивация, вызванная ДНК-метилированием, что также приводит к сильному падению продукции целевых белков при длительном культивировании клеток [109, 110]. Ретровирусные векторы также подвержены ДНК-метилированию в эмбриональных стволовых клетках, что приводит к мозаичной активности трансгенов в дочерних популяциях клеток [111, 112]. Помимо этого, системы доставки, обеспечивающие интеграцию трансгена в случайные районы генома, подвержены эффекту положения, в результате которого локальное воздействие хроматина непредсказуемым образом влияет на экспрессию [113].
Оптимизация системы экспрессии является сложным и трудозатратным процессом, которая включает в себя подбор системы доставки трансгена, отработку условий культивирования клеток-продуцентов и пр. [114]. Вместе с тем одним из основополагающих подходов для оптимизации системы экспрессии все же является рациональный дизайн генетической конструкции, проведенный с учетом типа клеток-продуцентов, а также требований к генотоксичности и уровню экспрессии.
Действительно, наличие в составе конструкции регуляторных элементов, которые могут оказывать влияние на продукцию трансгена как на транскрипционном, так и посттранскрипционном уровне, предоставляют массу возможностей для ее точной настройки. Например, добавление энхансеров или подбор подходящего промотора позволяет не только усилить транскрипцию, но также добиться тканеспеспецифичной активности трансгенов в отдельных клеточных популяциях организма, снижая потребность в использовании сложных систем таргетной доставки трансгенов для генной терапии [115–117]. Оптимизация последовательностей некодирующих областей транскриптов оказывает значительный эффект на стабильность мРНК и эффективность трансляции, что также позволяет повысить выход продукта [118–120].
Стабильность экспрессии трансгена также может быть повышена за счет противодействия эпигенетическим механизмам, оказывающим влияние на уровень экспрессии. Использование промоторов, не содержащих CpG-сайтов, позволяет исключить возможность метилирования ДНК и добиться стабильной экспрессии при длительном культивировании [121, 122]. Распространенным способом защиты от локального воздействия хроматиновых факторов является комплектация генетической конструкции регуляторными элементами, которые изолируют экспрессионную кассету. Такими элементами могут быть районы, ассоциированные с ядерным матриксом (SAR/MARs) [123, 124], районы контроля локуса (LCRs) [125], инсуляторы [126]

кт ГА
или элементы открытого хроматина (UCOEs) [127]. Помимо защиты от локального воздействия хроматина, использование таких элементов позволяет избежать генотоксичности трансгенной конструкции, возникающей, например, за счет аберрантной активации близлежащих генов ее регуляторными элементами [128, 129]. Дополняющим подходом является интеграция экспрессирующей конструкции в так называемые безопасные локусы генома методами направленного мутагенеза [130]. Интеграция в такие локусы обеспечивает предсказуемый уровень экспрессии и минимальную генотоксичность [131]. На настоящий момент в геноме человека охарактеризованы отдельные подобные локусы, что наряду с необходимостью применения систем направленной геномной интеграции трансгенов несколько ограничивает применимость таких подходов [132].
Создание экспрессионной конструкции невозможно без подбора оптимальных регуляторных последовательностей, что, хотя и является трудозатратным и длительным процессом, но зачастую позволяет кратно увеличить эффективность транскрипции [133, 134]. В этом контексте МПРА представляет собой удобный инструмент скрининга потенциальных регуляторных элементов, в том числе обеспечивающих заданный уровень активности [86]. Действительно, в работе [91] было продемонстрировано, что различные комбинации из тысячи промоторов и тысячи энхансеров позволяют достичь уровни транскрипции в широком диапазоне значений, охватывающем пять порядков. Несомненным преимуществом МПРА является скорость проведения анализа. Так, в работе [135] применение МПРА позволило определить энхансеры в бета-глобино-вом локусе человека и, в конечном итоге, за считанные недели значительно повысить эффектив- ность терапевтического лентивирусного вектора, экспрессирующего бета-глобин. Помимо поиска энхансеров и других ЦРЭ, разные вариации МПРА позволяют находить последовательности, которые могут увеличить выход продукта за счет оптимальной посттранскрипционной регуляции [101].
Методы МПРА могут быть использованы также для дизайна синтетических регуляторных элементов [136]. Так, тестирование тысяч последовательностей, различающихся единичными заменами, позволяет на основе количественных термодинамических моделей спрогнозировать структуру последовательностей, обладающих оптимальной активностью, а также экспериментально выявлять фрагменты ДНК с требуемыми свойствами [136–138].
Использование МПРА возможно также в фармакологии, поскольку применяется в том числе для поиска ЦРЭ, активирующихся под действием гормонов или химических веществ [139, 140]. Кроме того, МПРА был использован для оптимизации стабильности мРНК, что является одной из ключевых сложностей при применении мРНК-вакцин [141]. Разработанная недавно платформа МПРА под названием PERSIST-seq позволяет идентифицировать варианты последовательностей и элементы структуры транскрипта, которые позволяют значительно увеличить стабильность мРНК в растворах или средах организма [142].
Как уже упоминалось ранее, регуляторные элементы генома обеспечивают тканеспецифичные паттерны экспрессии, причем один и тот же участок геномной ДНК может проявлять дифференциальную активность в разных типах клеток или на различных стадиях развития [143, 144]. Методы МПРА неоднократно применялись для изучения тканеспецифичной функции регуляторных элементов [145–147]. Например, исследование в клетках глиобластомы U87 и нейтральных клетках-предшественниках позволило установить, что тканеспецифичные энхансеры обогащены мотивами связывания тех транскрипционных факторов, которые экспрессируются в данном клеточном типе [146].
Методом МПРА выявлены также промоторы, активность которых в клетках рака молочной железы была увеличена в 500 раз по сравнению с нормой, что позволяет потенциально применять их для терапевтических потребностей [137]. Таким образом, МПРА может помочь в создании систем тканеспецифичной экспрессии, что позволит минимизировать побочные эффекты генной терапии, связанные с нецелевой активностью трансгенов.
Применение массового параллельного репортерного анализа для идентификации клинически значимых мутаций
Методы полногеномного поиска ассоциаций (GWAS) позволяют идентифицировать геномные полиморфизмы, ассоциированные с различными патологиями [148–151]. Высокая частота встречаемости клинически значимых полиморфизмов в ЦРЭ позволяет предположить их влияние на регуляцию генов, задействованных в патогенезе заболевания [152]. Исследование транскриптома и GWAS-анализ позволяют предсказывать геномные локусы количественных признаков экспрессии (eQTL), ассоциированные с изменением уровня активности генов [153]. Благодаря этим методам созданы обширные базы данных, которые содержат информацию о генетических полиморфизмах, связанных с изменением активности генов, в том числе ассоциированных с заболеваниями человека [10]. Однако лишь незначительная часть информации, накопленной в базах данных, была подвергнута валидации в экспериментальных моделях [154], что делает затруднительным создание методов молекулярной диагностики и исследование механизмов, лежащих в основе заболеваний.
Методы МПРА, в свою очередь, позволяют валидировать функцию и уточнять клиническую значимость геномных полиморфизмов. Например, МПРА-скрининг 1605 однонуклеотидных полиморфизмов, ассоциированных с остеоартритом, согласно GWAS, позволил обнаружить связь между остеоартритом и нарушением сигнального пути Wnt в клетках [155]. Методом МПРА была проведена проверка регуляторного потенциала 1083 полиморфизмов из 73 некодирующих районов, ассоциированных с шизофренией и болезнью Альцгеймера, и выявлено 192 потенциально значимых для патогенеза полиморфизма [156]. Кроме того, исследование 5706 GWAS-полиморфизмов при помощи МПРА позволило обнаружить пять генов риска для синдрома Альцгеймера и прогрессирующего супрануклеарного пареза [157]. Методом МПРА выявлена также ключевая роль повышенной активности гена MX2 в патогенезе меланомы [158].
Вместе с тем анализ eQTL и МПРА являются, скорее, взаимодополняющими, поскольку МПРА, по всей вероятности, позволяет идентифицировать редкие функциональные регуляторные полиморфизмы, определение которых в анализе eQTL зачастую невозможно ввиду ограничений чувствительности метода [159]. В свою очередь МПРА также может давать несколько искаженную картину, поскольку экспериментальная проверка проводится in vitro в доступных клеточных культурах, далеко не всегда воспроизводящих физиологию клеток, задействованных в патогенезе заболевания, а также в основном сфокусирован на исследовании функции энхансеров [159].
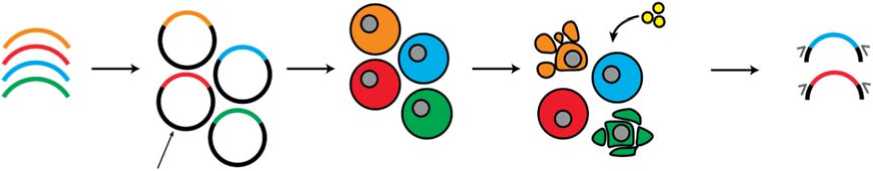
Совпадение результатов МПРА и eQTL имеет место приблизительно в 1/3 случаев, что, однако, вселяет надежду на высокую прогностическую ценность МПРА-позитивных eQTL локусов [159]. МПРА также дополняет исследования методом GWAS, поскольку, в отличие от него, позволяет сравнивать регуляторный эффект индивидуальных мутаций, давая возможность находить наиболее значимые полиморфизмы среди множества близкорасположенных сцепленных вариантов (рис. 2) [160]. Так, в исследовании мутаций с предсказанной эритроидной активностью МПРА позволил выявить 32 причинных варианта среди 2800 полиморфизмов, связанных сильным неравновесием по сцеплению [161]. А в исследовании аллельных вариантов, ассоциированных с риском диабета второго типа, МПРА позволил сузить круг предсказанных причинных вариантов с 52 до 1 [162].
Модификации МПРА, позволяющие исследовать мутации некодирующих областей гена, оказывающих влияние на стабильность мРНК и эффективность трансляции, также значительно расширяют возможности изучения патогенеза заболеваний, ассоциированных с такими нарушениями. Подобные нарушения обнаружены, например, при семейной дизавтономии — дегенеративном заболевании нервной системы, которое вызывается мутацией 5’-сайта сплайсинга в интроне гена IKBKAP , что приводит к исключению одного из экзонов и снижению уровня белка IKAP в нервной ткани [163]. Соматические мутации в 5’-нетранслируемых областях генов, которые потенциально влияют на эффективность трансляции, также обнаруживаются в клетках рака [164, 165]. Использование МПРА позволило идентифицировать мутации в некодирующих областях генов, ассоциированных с раком простаты [166], болезнями аутистического спектра [167], а также с другими патологическими состояниями и признаками человека [98, 168].
ЗАКЛЮЧЕНИЕ
Наличие обширных массивов данных, описывающих механизмы генетической и эпигенетической регуляции генов в норме и при патологии, а также внедрение функциональных методов геномного анализа позволяют значительно продвинуться в понимании механизмов точной регуляции экспрессии генов. Прогресс в этой области позволяет надеяться на возможность разработки количественных биологических моделей регуляции генной экспрессии, что, несомненно, придаст дополнительный стимул развитию подходов клинической и практической биотехнологии.
ДОПОЛНИТЕЛЬНАЯ ИНФОРМАЦИЯ
Вклад авторов. Авторы внесли равный вклад в создание статьи и подтверждают соответствие
FATAT
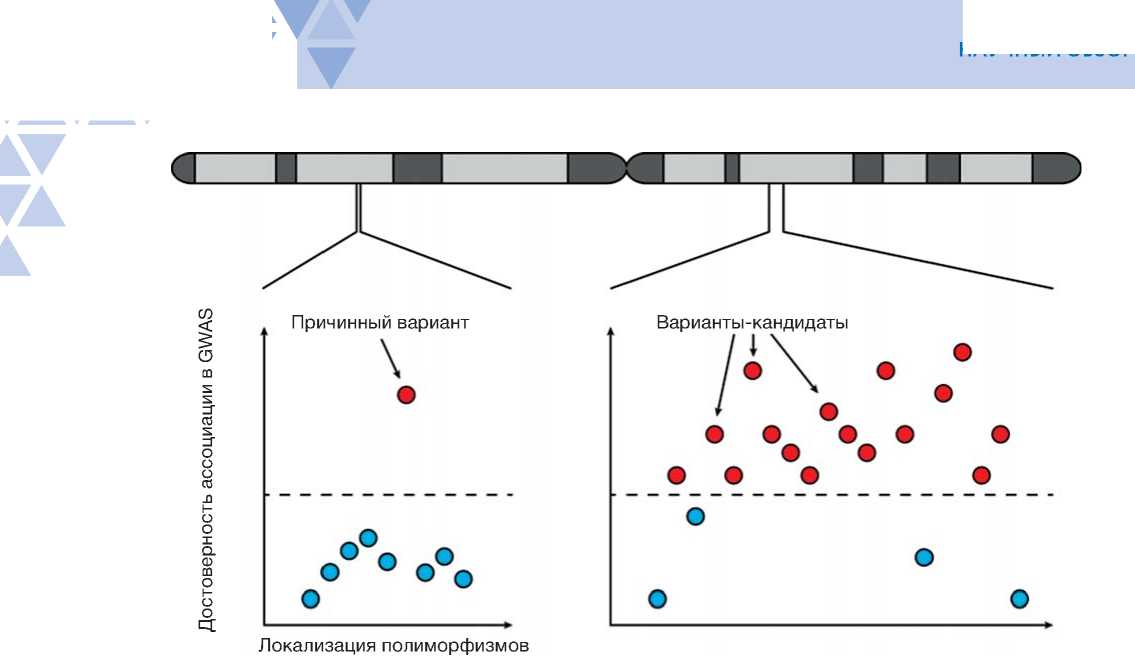
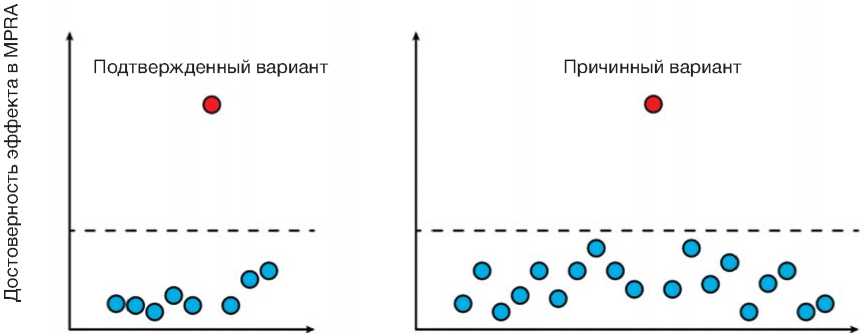
Рис. 2. Массовый параллельный репортерный анализ (МПРА) позволяет подтверждать причинные варианты среди множества кандидатов, идентифицированных при помощи GWAS (локус слева). В то же время мутации, вызывающие заболевание, часто входят в состав гаплотипов, включающих множество близко лежащих полиморфизмов (локус справа). В таких локусах GWAS не способен идентифицировать причинные генетические варианты, поэтому механизм развития болезни остается неясным. МПРА позволяет находить в гаплотипах полиморфизмы, нарушающие регуляцию генов, и выявлять молекулярные основы формирования болезни.
Fig. 2. Massively parallel reporter assays (MPRA) allow to confirms causative variants among the multiple candidates identified using GWAS (locus on the left). At the same time, causal mutations are often found within haplotypes containing multiple closely spaced polymorphisms (locus on the right). At such loci, GWAS is unable to identify the causal genetic variants, so the mechanism of disease development remains unclear. MPRA makes it possible to find polymorphisms in the haplotypes that disrupt gene regulation and identify the molecular basis of disease formation.
своего авторства международным критериям ICMJE (все авторы внесли существенный вклад в разработку концепции, проведение исследования и подготовку статьи, прочли и одобрили финальную версию перед публикацией).
Author contribution. The authors contributed equally to the article. The authors made a substantial contribution to the conception of the work, acquisition, analysis, interpretation of data for the work, drafting and revising the work, final approval of the version to be published and agree to be accountable for all aspects of the work.
Источник финансирования. Работа выполнена при финансовой поддержке Министерства науки и высшего образования Российской Федерации (Соглашение № 075-15-2021-1086, контракт № RF----193021X0015, 15.ИП.21.0015).
эактика
Funding source. This work was financially supported by the Ministry of Science and Higher Education of the Russian Federation (Agreement № 075-15-2021-1086, contract № RF----193021X0015, 15.ИП.21.0015).
Список литературы Практическое применение массового параллельного репортерного анализа в биотехнологии и медицине
- Oudelaar AM, Higgs DR. The relationship between genome structure and function. Nat Rev Genet. 2021;22(3):154–168. doi: 10.1038/s41576-020-00303-x
- Preissl S, Gaulton KJ, Ren B. Characterizing cis-regulatory elements using single-cell epigenomics. Nat Rev Genet. 2023; 24(1):21–43. doi: 10.1038/s41576-022-00509-1
- Bradner JE, Hnisz D, Young RA. Transcriptional addiction in cancer. Cell. 2017;168(4):629–643. doi: 10.1016/j.cell.2016.12.013
- Chatterjee S, Ahituv N. Gene regulatory elements, major drivers of human disease. Annu Rev Genomics Hum Genet. 2017;18: 45–63. doi: 10.1146/annurev-genom-091416-035537
- Corradin O, Saiakhova A, Akhtar-Zaidi B, et al. Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genom Res. 2014;24(1):1–13. doi: 10.1101/gr.164079.113
- Maurano MT, Humbert R. Rynes E, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337(6099):1190–1195. doi: 10.1126/science.1222794
- Claringbould A, Zaugg JB. Enhancers in disease: molecular basis and emerging treatment strategies. Trends Mol Med. 2021; 27(11):1060–1073. doi: 10.1016/j.molmed.2021.07.012
- Ernst J, Kheradpour P, Mikkelsen TS, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473(7345):43–49. doi: 10.1038/nature09906
- ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414): 57–74. doi: 10.1038/nature11247
- Lonsdale J, Thomas J, Salvatore M, et al. The genotype-tissue expression (GTEx) project. Nat Genet. 2013;45(6):580–585. doi: 10.1038/ng.2653
- Lee CP, Ko AM, Chiang SL, et al. Regulatory elements in vectors containing the ctEF-1α first intron and double enhancers for an efficient recombinant protein expression system. Scie Rep. 2018;8(1):15396. doi: 10.1038/s41598-018-33500-0
- Ingusci S, Verlengia G, Soukupova M, et al. Gene therapy tools for brain diseases. Front Pharmacol. 2019;10:724. doi: 10.3389/fphar.2019.00724
- Pan D, Büning H, Ling C. Rational design of gene therapy vectors. Mol Ther Method Clin Dev. 2019;12:246–247. doi: 10.1016/j.omtm.2019.01.009
- Xu D, Wang X, Jia Y, et al. SV40 intron, a potent strong intron element that effectively increases transgene expression in transfected Chinese hamster ovary cells. J Cell Mol Med. 2018; 22(4):2231–2239. doi: 10.1111/jcmm.13504
- Romanov SE, Kalashnikova DA, Laktionov PP. Methods of massive parallel reporter assays for investigation of enhancers. Vavilovskii Zhurnal Genet Selektsii. 2021;25(3):344–355. doi: 10.18699/VJ21.038
- Thurman RE, Rynes E, Humbert R, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489(7414):75–82. doi: 10.1038/nature11232
- Andersson R, Sandelin A. Determinants of enhancer and promoter activities of regulatory elements. Nat Rev Genet. 2020;21(2):71–87. doi: 10.1038/s41576-019-0173-8
- Furlong EE, Levine M. Developmental enhancers and chromosome topology. Science. 2018;361(6409):1341–1345. doi: 10.1126/science.aau0320
- Pang B, van Weerd JH, Hamoen FL, Snyder MP. Identification of non-coding silencer elements and their regulation of gene expression. Nat Rev Mol Cell Biol. 2022. doi: 10.1038/s41580-022-00549-9
- Chen D, Lei EP. Function and regulation of chromatin insulators in dynamic genome organization. Curr Opin Cell Biol. 2019;58: 61–68. doi: 10.1016/j.ceb.2019.02.001
- Haberle V, Stark A. Eukaryotic core promoters and the functional basis of transcription initiation. Nat Rev Mol Cell Biol. 2018;19(10):621–637. doi: 10.1038/s41580-018-0028-8
- Lettice LA, Williamson I, Devenney PS, et al. Development of five digits is controlled by a bipartite long-range cis-regulator. Development. 2014;141(8):1715–1725. doi: 10.1242/dev.095430
- Pennacchio LA, Bickmore W, Dean A, et al. Enhancers: five essential questions. Nat Rev Gen. 2013;14(4):288–295. doi: 10.1038/nrg3458
- Dickel DE, Ypsilanti AR, Pla R, et al. Ultraconserved enhancers are required for normal development. Cell. 2018;172(3):491–499. e15. doi: 10.1016/j.cell.2017.12.017
- Sur I, Taipale J. The role of enhancers in cancer. Nat Rev Canc. 2016;16(8):483–493. doi: 10.1038/nrc.2016.62
- Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98(3):387–396. doi: 10.1016/s0092-8674(00)81967-4
- Kellum R, Schedl P. A position-effect assay for boundaries of higher order chromosomal domains. Cell. 1991;64(5):941–950. doi: 10.1016/0092-8674(91)90318-s
- Davidson IF, Peters JM. Genome folding through loop extrusion by SMC complexes. Nat Rev Mol Cell Biol. 2021;22(7):445–464. doi: 10.1038/s41580-021-00349-7
- Hnisz D, Day DS, Young RA. Insulated neighborhoods: structural and functional units of mammalian gene control. Cell. 2016;167(5):1188–1200. doi: 10.1016/j.cell.2016.10.024
- Dixon JR, Selvaraj S, Yue F, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485(7398):376–380. doi: 10.1038/nature11082
- Flavahan WA, Drier Y, Johnstone SE, et al. Altered chromosomal topology drives oncogenic programs in SDH-deficient GISTs. Nature. 2019;575(7781):229–233. doi: 10.1038/s41586-019-1668-3
- Hnisz D, Weintraub AS, Day DS, et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science. 2016;351(6280):1454–1458. doi: 10.1126/science.aad9024
- Rhie SK, Perez AA, Lay FD, et al. A high-resolution 3D epigenomic map reveals insights into the creation of the prostate cancer transcriptome. Nat Commun. 2019;10(1):4154. doi: 10.1038/s41467-019-12079-8
- Sun JH, Zhou L, Emerson DJ, et al. Disease-associated short tandem repeats co-localize with chromatin domain boundaries. Cell. 2018;175(1):224–238.e15. doi: 10.1016/j.cell.2018.08.005
- Harris MB, Mostecki J, Rothman PB. Repression of an interleukin-4- responsive promoter requires cooperative BCL-6 function. J Biol Chem. 2005;280(13):13114–13121. doi: 10.1074/jbc.M412649200
- Lanzuolo C, Roure V, Dekker J, et al. Polycomb response elements mediate the formation of chromosome higher-order structures in the bithorax complex. Nat Cell Biol. 2007;9(10): 1167–1174. doi: 10.1038/ncb1637
- Mori N, Stein R, Sigmund O, Anderson DJ. A cell type-preferred silencer element that controls the neural-specific expression of the SCG10 gene. Neuron. 1990;4(4):583–594. doi: 10.1016/0896-6273(90)90116-w
- Li L, He S, Sun JM, Davie JR. Gene regulation by Sp1 and Sp3. Biochem Cell Biol. 2004;82(4):460–471. doi: 10.1139/o04-045
- Srinivasan L, Atchison ML. YY1 DNA binding and PcG recruitment requires CtBP. Genes Dev. 2004;18(21):2596–2601. doi: 10.1101/gad.1228204
- Tiwari VK, McGarvey KM, Licchesi JD, et al. PcG Proteins, DNA methylation, and gene repression by chromatin looping. PLoS Biol. 2008;6(12):e306. doi: 10.1371/journal.pbio.0060306
- Segert JA, Gisselbrecht SS. Bulyk ML. Transcriptional silencers: driving gene expression with the brakes on. Trends Genet. 2021;37(6):514–527. doi: 10.1016/j.tig.2021.02.002
- Andersson R, Gebhard C, Miguel-Escalada I, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507(7493):455–461. doi: 10.1038/nature12787
- Bernstein BE, Stamatoyannopoulos JA, Costello JF, et al. The NIH roadmap epigenomics mapping consortium. Nat Biotechnol. 2010;28(10):1045–1048. doi: 10.1038/nbt1010-1045
- ENCODE Project Consortium; Birney E, Stamatoyannopoulos JA, Dutta A, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816. doi: 10.1038/nature05874
- FANTOM Consortium and the RIKEN PMI and CLST (DGT); Forrest AR, Kawaji H, Rehli M, et al. A promoter-level mammalian expression atlas. Nature. 2014;507(7493):462–470. doi: 10.1038/nature13182
- Kellis M, Wold B, Snyder MP, et al. Defining functional DNA elements in the human genome. Proc Natl Acad Sci USA. 2014;111(17):6131–6138. doi: 10.1073/pnas.1318948111
- Roadmap Epigenomics Consortium; Kundaje A, Meuleman W, Ernst J, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518(7539):317–330. doi: 10.1038/nature14248
- Bernstein BE, Kamal M, Lindblad-Toh K, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120(2):169–181. doi: 10.1016/j.cell.2005.01.001
- Spicuglia S, Vanhille L. Chromatin signatures of active enhancers. Nucleus. 2012;3(2):126–131. doi: 10.4161/nucl.19232
- Visel A, Blow MJ, Li Z, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457(7231): 854–858. doi: 10.1038/nature07730
- Rada-Iglesias A, Bajpai R, Swigut T, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470(7333):279–283. doi: 10.1038/nature09692
- Whyte WA, Orlando DA, Hnisz D, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153(2):307–319. doi: 10.1016/j.cell.2013.03.035
- Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007; 129(4):823–837. doi: 10.1016/j.cell.2007.05.009
- Pang B, Snyder MP. Systematic identification of silencers in human cells. Nat Genet. 2020;52(3):254–263. doi: 10.1038/s41588-020-0578-5
- Dixon JR, Jung I, Selvaraj S, et al. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015; 518(7539):331–336. doi: 10.1038/nature14222
- Nora EP, Goloborodko A, Valton AL, et al. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell. 2017;169(5): 930–944.e22. doi: 10.1016/j.cell.2017.05.004
- Rao SS, Huang SC, Glenn St Hilaire B, et al. Cohesin loss eliminates all loop domains. Cell. 2017;171(2):305–320.e24. doi: 10.1016/j.cell.2017.09.026
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007; 316(5830):1497–1502. doi: 10.1126/science.1141319
- Rhee HS, Pugh BF. Comprehensive genome-wide protein- DNA interactions detected at single-nucleotide resolution. Cell. 2011;147(6):1408–1419. doi: 10.1016/j.cell.2011.11.013
- John S, Sabo PJ, Thurman RE, et al. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet. 2011;43(3):264–268. doi: 10.1038/ng.759
- Doni Jayavelu N, Jajodia A, Mishra A, Hawkins RD. Candidate silencer elements for the human and mouse genomes. Nat Commun. 2020;11(1):1061. doi: 10.1038/s41467-020-14853-5
- Xu B, Wang H, Wright S, et al. Acute depletion of CTCF rewires genome-wide chromatin accessibility. Genome Biol. 2021;22(1):244. doi: 10.1186/s13059-021-02466-0
- Song L, Zhang Z, Grasfeder LL, et al. Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res. 2011;21(10):1757–1767. doi: 10.1101/gr.121541.111
- Valouev A, Johnson SM, Boyd SD, et al. Determinants of nucleosome organization in primary human cells. Nature. 2011; 474(7352):516–520. doi: 10.1038/nature10002
- Song L, Crawford GE. DNase-seq: a high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harb Protoc. 2010; 2010(2):pdb.prot5384. doi: 10.1101/pdb.prot5384
- Pajoro A, Muiño JM, Angenent GC, Kaufmann K. Profiling nucleosome occupancy by MNase-seq: experimental protocol and computational analysis. Methods Mol Biol. 2018;1675: 167–181. doi: 10.1007/978-1-4939-7318-7_11
- Buenrostro JD, Giresi PG, Zaba LC, et al. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10(12):1213–1218. doi: 10.1038/nmeth.2688
- Giresi PG, Kim J, McDaniell RM, et al. FAIRE (Formaldehyde- Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007; 17(6):877–885. doi: 10.1101/gr.5533506
- Lu L, Liu X, Huang WK, et al. Robust Hi-C maps of enhancerpromoter interactions reveal the function of non-coding genome in neural development and diseases. Mol Cell. 2020;79(3): 521–534.e15. doi: 10.1016/j.molcel.2020.06.007
- Ngan CY, Wong CH, Tjong H, et al. Chromatin interac tion analyses elucidate the roles of PRC2-bound silencers in mouse development. Nat Genet. 2020;52(3):264–272. doi: 10.1038/s41588-020-0581-x
- Sun F, Chronis C, Kronenberg M, et al. Promoterenhancer communication occurs primarily within insulated neighborhoods. Mol Cell. 2019;73(2):250–263.e5. doi: 10.1016/j.molcel.2018.10.039
- Li Z, McGinn O, Wu Y, et al. ESR1 mutant breast cancers show elevated basal cytokeratins and immune activation. Nat Commun. 2022;13(1):2011. doi: 10.1038/s41467-022-29498-9
- Lieberman-Aiden E, van Berkum NL, Williams L, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326(5950): 289–293. doi: 10.1126/science.1181369
- Fullwood MJ, Ruan Y. ChIP-based methods for the identification of long-range chromatin interactions. J Cell Biochem. 2009; 107(1):30–39. doi: 10.1002/jcb.22116
- Mumbach MR, Rubin AJ, Flynn RA, et al. HiChIP: Efficient and sensitive analysis of protein-directed genome architecture. Nat Methods. 2016;13(11):919–922. doi: 10.1038/nmeth.3999
- Fang R, Yu M, Li G, et al. Mapping of long-range chromatin interactions by proximity ligation-assisted ChIP-seq. Cell Res. 2016;26(12):1345–1348. doi: 10.1038/cr.2016.137
- Banerji J, Rusconi S, Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27(2 Pt 1):299–308. doi: 10.1016/0092-8674(81)90413-x
- Kvon EZ, Kazmar T, Stampfel G, et al. Genome-scale functional characterization of Drosophila developmental enhancers in vivo. Nature. 2014;512(7512):91–95. doi: 10.1038/nature13395
- Patwardhan RP, Lee C, Litvin O, et al. High-resolution analysis of DNA regulatory elements by synthetic saturation mutagenesis. Nat Biotechnol. 2009;27(12):1173–1175. doi: 10.1038/nbt.1589
- Patwardhan RP, Hiatt JB, Witten DM, et al. Massively parallel functional dissection of mammalian enhancers in vivo. Nat Biotechnol. 2012;30(3):265–270. doi: 10.1038/nbt.2136
- Melnikov A, Murugan A, Zhang X, et al. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat Biotechnol. 2012;30(3): 271–277. doi: 10.1038/nbt.2137
- Kwasnieski JC, Mogno I, Myers CA, et al. Complex effects of nucleotide variants in a mammalian cis-regulatory element. Proc Natl Acad Sci USA. 2012;109(47):19498–19503. doi: 10.1073/pnas.1210678109
- McAfee JC, Bell JL, Krupa O, et al. Focus on your locus with a massively parallel reporter assay. J Neurodev Disord. 2022; 14(1):50. doi: 10.1186/s11689-022-09461-x
- Kheradpour P, Ernst J, Melnikov A, et al. Systematic dissection of regulatory motifs in 2000 predicted human enhancers using a massively parallel reporter assay. Genome Res. 2013;23(5): 800–811. doi: 10.1101/gr.144899.112
- Kwasnieski JC, Fiore C, Chaudhari HG, Cohen BA. Highthroughput functional testing of ENCODE segmentation predictions. Genome Res. 2014;24(10):1595–1602. doi: 10.1101/gr.173518.114
- Arnold CD, Gerlach D, Stelzer C, et al. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science. 2013; 339(6123):1074–1077. doi: 10.1126/science.1232542
- Nguyen TA, Jones RD, Snavely AR, et al. High-throughput functional comparison of promoter and enhancer activities. Genome Res. 2016;26(8):1023–1033. doi: 10.1101/gr.204834.116
- Hansen TJ, Hodges E. ATAC-STARR-seq reveals transcription factor-bound activators and silencers within chromatinaccessible regions of the human genome. Genome Res. 2022; 32(8):1529–1541. doi: 10.1101/gr.276766.122
- Pang B, Neijssen J, Qiao X, et al. Direct antigen presentation and gap junction mediated cross-presentation during apoptosis. J Immunol. 2009;183(2):1083–1090. doi: 10.4049/jimmunol.0900861
- Straathof KC, Pulè MA, Yotnda P, et al. An inducible caspase 9 safety switch for T-cell therapy. Blood. 2005;105(11):4247–4254. doi: 10.1182/blood-2004-11-4564
- Bergman DT, Jones TR, Liu V, et al. Compatibility rules of human enhancer and promoter sequences. Nature. 2022;607(7917): 176–184. doi: 10.1038/s41586-022-04877-w
- Davis JE, Insigne KD, Jones EM, et al. Dissection of c-AMP response element architecture by using genomic and episomal massively parallel reporter assays. Cell Syst. 2020;11(1):75–85. e7. doi: 10.1016/j.cels.2020.05.011
- Ribeiro-dos-Santos AM, Hogan MS, Luther RD, et al. Genomic context sensitivity of insulator function. Genome Res. 2022; 32(3):425–436. doi: 10.1101/gr.276449.121
- Rosenberg AB, Patwardhan RP, Shendure J, Seelig G. Learning the sequence determinants of alternative splicing from millions of random sequences. Cell. 2015;163(3):698–711. doi: 10.1016/j.cell.2015.09.054
- Cheung R, Insigne KD, Yao D, et al. A multiplexed assay for exon recognition reveals that an unappreciated fraction of rare genetic variants cause large-effect splicing disruptions. Mol Cell. 2019;73(1):183–194.e8. doi: 10.1016/j.molcel.2018.10.037
- Mikl M, Hamburg A, Pilpel Y, Segal E. Dissecting splicing decisions and cell-to-cell variability with designed sequence libraries. Nat Commun. 2019;10(1):4572. doi: 10.1038/s41467-019-12642-3
- Matreyek KA, Starita LM, Stephany JJ, et al. Multiplex assessment of protein variant abundance by massively parallel sequencing. Nat Genet. 2018;50(6):874–882. doi: 10.1038/s41588-018-0122-z
- Sample PJ, Wang B, Reid DW, et al. Human 5' UTR design and variant effect prediction from a massively parallel translation assay. Nat Biotechnol. 2019;37(7):803–809. doi: 10.1038/s41587-019-0164-5
- Vainberg Slutskin I, Weinberger A, Segal E. Sequence determinants of polyadenylation-mediated regulation. Genome Res. 2019;29(10):1635–1647. doi: 10.1101/gr.247312.118
- Safra M, Nir R, Farouq D, et al. TRUB1 is the predominant pseudouridine synthase acting on mammalian mRNA via a predictable and conserved code. Genome Res. 2017;27(3): 393–406. doi: 10.1101/gr.207613.116
- Mulvey B, Lagunas T, Dougherty JD. Massively parallel reporter assays: defining functional psychiatric genetic variants across biological contexts. Biol Psych. 2021;89(1): 6–89. doi: 10.1016/j.biopsych.2020.06.011
- Zopf CJ, Quinn K, Zeidman J, Maheshri N. Cell-cycle dependence of transcription dominates noise in gene expression. PLoS Comput Biol. 2013;9(7):e1003161. doi: 10.1371/journal.pcbi.1003161
- Akita H, Ito R. Kamiya H, et al. Cell cycle dependent transcription, a determinant factor of heterogeneity in cationic lipid-mediated transgene expression. J Gene Med. 2007;9(3):197–207. doi: 10.1002/jgm.1010
- Cooper LJ, Topp MS, Pinzon C, et al. Enhanced transgene expression in quiescent and activated human CD8 + T cells. Human Gene Therapy. 2004;15(7):648–658. doi: 10.1089/1043034041361217
- Brightwell G, Poirier V, Cole E, et al. Serum-dependent and cell cycle-dependent expression from a cytomegalovirusbased mammalian expression vector. Gene. 1997;194(1): 115–123. doi: 10.1016/s0378-1119(97)00178-9
- Dutton RL, Scharer J, Moo-Young M. Cell cycle phase dependent productivity of a recombinant Chinese hamster ovary cell line. Cytotechnology. 2006;52(1):55–69. doi: 10.1007/s10616-006-9041-4
- Burns WR, Zheng Z, Rosenberg SA, Morgan RA. Lack of specific γ-retroviral vector long terminal repeat promoter silencing in patients receiving genetically engineered lymphocytes and activation upon lymphocyte restimulation. Blood. 2009; 114(14):2888–2899. doi: 10.1182/blood-2009-01-199216
- Rosenqvist N, Segerstad C, Samuelsson C, et al. Activation of silenced transgene expression in neural precursor cell lines by inhibitors of histone deacetylation. J Gene Med. 2002; 4(3):248–257. doi: 10.1002/jgm.268
- Kim M, O’Callaghan PM, Droms KA, James DC. A mechanistic understanding of production instability in CHO cell lines expressing recombinant monoclonal antibodies. Biotechnol Bioeng. 2011;108(10):2434–2446. doi: 10.1002/bit.23189
- Brooks AR, Harkins RN, Wang P, et al. Transcriptional silencing is associated with extensive methylation of the CMV promoter following adenoviral gene delivery to muscle. J Gene Med. 2004;6(4):395–404. doi: 10.1002/jgm.516
- Laker C, Meyer J, Schopen A, et al. Host cis-mediated extinction of a retrovirus permissive for expression in embryonal stem cells during differentiation. J Virol. 1998;72(1):339–348. doi: 10.1128/JVI.72.1.339-348.1998
- Yao S, Sukonnik T, Kean T, et al. Retrovirus silencing, variegation, extinction, and memory are controlled by a dynamic interplay of multiple epigenetic modifications. Mol Ther. 2004;10(1):27–36. doi: 10.1016/j.ymthe.2004.04.007
- Cranston A, Dong C, Howcroft J, Clark AJ. Chromosomal sequences flanking an efficiently expressed transgene dramatically enhance its expression. Gene. 2001;269(1–2): 217–225. doi: 10.1016/s0378-1119(01)00459-0
- Tripathi NK, Shrivastava A. Recent developments in bioprocessing of recombinant proteins: expression hosts and process development. Front Bioeng Biotechnol. 2019;7:420. doi: 10.3389/fbioe.2019.00420
- Nair RR, Blankvoort S, Lagartos MJ, Kentros C. Enhancerdriven gene expression (edge) enables the generation of viral vectors specific to neuronal subtypes. Science. 2020;23(3): 100888. doi: 10.1016/j.isci.2020.100888
- Gruh I, Wunderlich S, Winkler M, et al. Human CMV imme diateearly enhancer: a useful tool to enhance cell-type-specific expression from lentiviral vectors. J Gene Med. 2008;10(1): 21–32. doi: 10.1002/jgm.1122
- Montaño-Samaniego M, Bravo-Estupiñan DM, Méndez- Guerrero O, et al. Strategies for targeting gene therapy in cancer cells with tumor-specific promoters. Front Oncol. 2020;10:605380. doi: 10.3389/fonc.2020.605380
- Sano KI, Maeda K, Oki M, Maéda Y. Enhancement of protein expression in insect cells by a lobster tropomyosin cDNA leader sequence. FEBS Lett. 2002;532(1–2):143–146. doi: 10.1016/s0014-5793(02)03659-1
- Eisenhut P, Mebrahtu A, Moradi Barzadd M, et al. Systematic use of synthetic 5'-UTR RNA structures to tune protein translation improves yield and quality of complex proteins in mammalian cell factories. Nucleic Acids Res. 2020;48(20): e119–e119. doi: 10.1093/nar/gkaa847
- Peyret H, Brown JK, Lomonossoff GP. Improving plant transient expression through the rational design of synthetic 5' and 3' untranslated regions. Plant Methods. 2019;15(1):108. doi: 10.1186/s13007-019-0494-9
- Moritz B, Becker PB, Göpfert U. CMV promoter mutants with a reduced propensity to productivity loss in CHO cells. Sci Rep. 2015;5(1):16952. doi: 10.1038/srep16952
- Ho SC, Koh EY, Soo BP, et al. Evaluating the use of a CpG free promoter for long-term recombinant protein expression stability in Chinese hamster ovary cells. BMC Biotechnol. 2016;16(1):71. doi: 10.1186/s12896-016-0300-y
- Kim JM, Kim JS, Park DH, et al. Improved recombinant gene expression in CHO cells using matrix attachment regions. J Biotechnol. 2004;107(2):95–105. doi: 10.1016/j.jbiotec.2003.09.015
- Phi-Van L, von Kries JP, Ostertag W, Strätling WH. The chicken lysozyme 5’ matrix attachment region increases transcription from a heterologous promoter in heterologous cells and dampens position effects on the expression of transfected genes. Mol Cell Biol. 1990;10(5):2302–2307. doi: 10.1128/mcb.10.5.2302-2307.1990
- Grosveld F, van Assendelft GB, Greaves DR, Kollias G. Position-independent, high-level expression of the human β-globin gene in transgenic mice. Cell. 1987;51(6):975–985. doi: 10.1016/0092-8674(87)90584-8
- Pikaart MJ, Recillas-Targa F, Felsenfeld G. Loss of transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation and is prevented by insulators. Genes Dev.1998;12(18):2852–2862. doi: 10.1101/gad.12.18.2852
- Neville JJ, Orlando J, Mann K, et al. Ubiquitous chromatinope ning elements (UCOEs): Applications in biomanufactu ring and gene therapy. Biotechnol Adv. 2017;35(5):557–564. doi: 10.1016/j.biotechadv.2017.05.004
- Li CL, Xiong D, Stamatoyannopoulos G, Emery DW. Genomic and functional assays demonstrate reduced gammaretroviral vector genotoxicity associated with use of the cHS4 chromatin insulator. Mol Ther. 2009;17(4):716–724. doi: 10.1038/mt.2009.7
- Browning D, Trobridge G. Insulators to improve the safety of retroviral vectors for HIV gene therapy. Biomedicines. 2016;4(1):4. doi: 10.3390/biomedicines4010004
- Hayashi H, Kubo Y, Izumida M, Matsuyama T. Efficient viral delivery of Cas9 into human safe harbor. Sci Rep. 2020; 10(1):21474. doi: 10.1038/s41598-020-78450-8
- Papapetrou EP, Schambach A. Gene insertion into genomic safe harbors for human gene therapy. Mol Ther. 2016;24(4): 678–684. doi: 10.1038/mt.2016.38
- Sadelain M, Papapetrou EP, Bushman FD. Safe harbours for the integration of new DNA in the human genome. Nat Rev Canc. 2011;12(1):51–58. doi: 10.1038/nrc3179
- Wang W, Guo X, Li Y, et al. Enhanced transgene expression using cis-acting elements combined with the EF1 promoter in a mammalian expression system. Eur J Pharm Sci. 2018; 123:539–545. doi: 10.1016/j.ejps.2018.08.016
- Schlabach MR, Hu JK, Li M, Elledge SJ. Synthetic design of strong promoters. Proc Natl Acad Sci USA. 2010;107(6): 2538–2543. doi: 10.1073/pnas.0914803107
- Morgan RA, Ma F, Unti MJ, et al. Creating new β-globinexpressing lentiviral vectors by high-resolution mapping of locus control region enhancer sequences. Mol Ther Methods Clin Dev. 2020;17:999–1013. doi: 10.1016/j.omtm.2020.04.006
- Vaknin I, Amit R. Molecular and experimental tools to design synthetic enhancers. Curr Opin Biotechnol. 2022;76:102728. doi: 10.1016/j.copbio.2022.102728
- Wu MR, Nissim L, Stupp D, et al. A high-throughput screening and computation platform for identifying synthetic promoters with enhanced cell-state specificity (SPECS). Nat Commun. 2019;10(1):2880. doi: 10.1038/s41467-019-10912-8
- Yu TC, Liu WL, Brinck MS, et al. Multiplexed characterization of rationally designed promoter architectures deconstructs combinatorial logic for IPTG-inducible systems. Nat Commun. 2021;12(1):325. doi: 10.1038/s41467-020-20094-3
- Shlyueva D, Stelzer C, Gerlach D, et al. Hormone-responsive enhancer-activity maps reveal predictive motifs, indirect repression, and targeting of closed chromatin. Mol Cell. 2014; 54(1):180–192. doi: 10.1016/j.molcel.2014.02.026
- Johnson GD, Barrera A, McDowell IC, et al. Human genomewide measurement of drug-responsive regulatory activity. Nat Commun. 2018;9(1):5317. doi: 10.1038/s41467-018-07607-x
- Pardi N, Hogan MJ, Porter FW. Weissman D. mRNA vaccines--a new era in vaccinology. Nat Rev Drug Dis. 2018;17(4): 261–279. doi: 10.1038/nrd.2017.243
- Leppek K, Byeon GW, Kladwang W, et al. Combinatorial optimization of mRNA structure, stability, and translation for RNA-based therapeutics. Nat Commun. 2022;13(1):1536. doi: 10.1038/s41467-022-28776-w
- Peng T, Zhai Y, Atlasi Y, et al. STARR-seq identifies active, chromatin-masked, and dormant enhancers in pluripotent mouse embryonic stem cells. Genome Biol. 2020;21(1):243. doi: 10.1186/s13059-020-02156-3
- Glaser LV, Steiger M, Fuchs A, et al. Assessing genome-wide dynamic changes in enhancer activity during early mESC differentiation by FAIRE-STARR-seq. Nucleic Acids Res. 2021; 49(21):12178–12195. doi: 10.1093/nar/gkab1100
- Van Arensbergen J, FitzPatrick VD, de Haas M, et al. Genomewide mapping of autonomous promoter activity in human cells. Nat Biotechnol. 2017;35(2):145–153. doi: 10.1038/nbt.3754
- Maricque BB, Dougherty JD, Cohen BA. A genome-integrated massively parallel reporter assay reveals DNA sequence determinants of cis-regulatory activity in neural cells. Nucleic Acids Res. 2017;45(4):e16. doi: 10.1093/nar/gkw942
- Inoue F, Kreimer A, Ashuach T, et al. Identification and massively parallel characterization of regulatory elements driving neural induction. Cell Stem Cell. 2019;25(5):713–727.e10. doi: 10.1016/j.stem.2019.09.010
- Dekkers CM. Application of genomics tools to animal breeding. Curr Genom. 2012;13(3):207–212. doi: 10.2174/138920212800543057
- Xiao Y, Liu H, Wu L, et al. Genome-wide association studies in maize: praise and stargaze. Mol Plant. 2017;10(3):359–374. doi: 10.1016/j.molp.2016.12.008
- Mackay TF, Richards S, Stone EA, et al. The drosophila melanogaster genetic reference panel. Nature. 2012;482(7384): 173–178. doi: 10.1038/nature10811
- Husby A, Kawakami T, Rönnegаrd L, et al. Genome-wide association mapping in a wild avian population identifies a link between genetic and phenotypic variation in a lifehistory trait. Proc Biol Sci. 2015;282(1806):20150156. doi: 10.1098/rspb.2015.0156
- Tseng CC, Wong MC, Liao WT, et al. Genetic variants in transcription factor binding sites in humans: triggered by natural selection and triggers of diseases. Int J Mol Sci. 2021; 22(8):4187. doi: 10.3390/ijms22084187
- Jung S, Liu W, Baek J, et al. Expression quantitative trait loci (eQTL) mapping in korean patients with crohn’s disease and identification of potential causal genes through integration with disease associations. Front Genet. 2020;11:486. doi: 10.3389/fgene.2020.00486
- Alsheikh AJ, Wollenhaupt S, King EA, et al. The landscape of GWAS validation; systematic review identifying 309 validated non-coding variants across 130 human diseases. BMC Med Genomics. 2022;15(1):74. doi: 10.1186/s12920-022-01216-w
- Klein JC, Keith A, Rice SJ, et al. Functional testing of thousands of osteoarthritis-associated variants for regulatory activity. Nat Commun. 2019;10(1):2434. doi: 10.1038/s41467-019-10439-y
- Myint L, Wang R, Boukas L, et al. A screen of 1,049 schizophrenia and 30 Alzheimer’s‐associated variants for regulatory potential. Am J Med Genet B Neuropsychiatr Genet. 2020; 183(1):61–73. doi: 10.1002/ajmg.b.32761
- Cooper YA, Teyssier N, Dräger NM, et al. Functional regulatory variants implicate distinct transcriptional networks in dementia. Science. 2022;377(6608):eabi8654. doi: 10.1126/science.abi8654
- Choi J, Zhang T, Vu A, et al. Massively parallel reporter assays of melanoma risk variants identify MX2 as a gene promoting melanoma. Nat Commun. 2020;11(1):2718. doi: 10.1038/s41467-020-16590-1
- McAfee JC, Lee S, Lee J, et al. Systematic investigation of alle lic regulatory activity of schizophrenia-associated com mon variants. medRxiv. 2022. Available from: https://www.researchgate.net/scientific-contributions/Jessica-Lee-Bell-2203854579
- Slatkin M. Linkage disequilibrium--understanding the evolutionary past and mapping the medical future. Nat Rev Gen. 2008;9(6):477–485. doi: 10.1038/nrg2361
- Ulirsch JC, Nandakumar SK, Wang L, et al. Systematic functional dissection of common genetic variation affecting red blood cell traits. Cell. 2016;165(6):1530–1545. doi: 10.1016/j.cell.2016.04.048
- Khetan S, Kales S, Kursawe R, et al. Functional characterization of T2D-associated SNP effects on baseline and ER stressresponsive β-cell transcriptional activation. Nat Commun. 2021;12(1):5242. doi: 10.1038/s41467-021-25514-6
- Dietrich P, Dragatsis I. Familial dysautonomia: mechanisms and models. Genet Mol Biol. 2016;39(4):497–514. doi: 10.1590/1678-4685-GMB-2015-0335
- Fujimoto A, Furuta M, Totoki Y, et al. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat Genet. 2016;48(5):500–509. doi: 10.1038/ng.3547
- Rheinbay E, Nielsen MM, Abascal F, et al. Analyses of noncoding somatic drivers in 2,658 cancer whole genomes. Nature. 2020;578(7793):102–111. doi: 10.1038/s41586-020-1969-6
- Lim Y, Arora S, Schuster SL, et al. Multiplexed functional genomic analysis of 5’ untranslated region mutations across the spectrum of prostate cancer. Nat Commun. 2021;12(1):4217. doi: 10.1038/s41467-021-24445-6
- Lagunas T, Plassmyer S, Friedman R, et al. A cre-dependent massively parallel reporter assay allows for cell-type specific assessment of the functional effects of genetic variants in vivo. bioRxiv. 2021. doi: 10.1101/2021.05.17.444514
- Griesemer D, Xue JR, Reilly SK, et al. Genome-wide functional screen of 3'UTR variants uncovers causal variants for human disease and evolution. Cell. 2021;184(20):5247–5260.e19. doi: 10.1016/j.cell.2021.08.025


