Prevalence of pan drug-resistant, extensively drug-resistant, and detection of multiple antibiotic resistance in uncommon bacterial isolates in Al-Al Zahraa teaching hospital for maternity and children in Najaf/Iraq
Автор: Ali Beg K.A.A., Khudair A.H., Hussein Etaij D.M., Mezher A.A., Aziz Ali beg A.A.
Журнал: Cardiometry @cardiometry
Рубрика: Original research
Статья в выпуске: 30, 2024 года.
Бесплатный доступ
Objectives: There are numerous opportunistic bacterial spe-cies that are unusual and rarely found in laboratory samples, most are challenging to routinely identify, some of them even being documented very little in clinical samples. Also, the role of these species in the coordinates of the disease is no less av-erage and important than the common bacterial species.
Pdr, xdr, mar, antibiotic resistance, uncommon bacteria
Короткий адрес: https://sciup.org/148328269
IDR: 148328269 | DOI: 10.18137/cardiometry.2024.30.4149
Текст научной статьи Prevalence of pan drug-resistant, extensively drug-resistant, and detection of multiple antibiotic resistance in uncommon bacterial isolates in Al-Al Zahraa teaching hospital for maternity and children in Najaf/Iraq
Karrar Abdil Aziz Ali Beg, Asmaa Hassan Khudair, Dafer Ma-hammad Hussein Etaij, Ahmed Abdullah Mezher, Ammar Abdul Aziz Ali beg. Prevalence of Pan drug-resistant, Extensively drug-resistant, and Detection of Multiple Antibiotic Resistance in Uncommon Bacterial Isolates in Al-Al Zahraa Teaching hospital for Maternity and Children in Najaf/Iraq. Cardiometry; Issue No. 30; February 2024; p. 41-49; DOI: 10.18137/cardiom-etry.2024.30.4149; Available from: issues/no30-february-2024/prevalence-pan-drug-resistant-ex-tensively
There are numerous opportunistic bacterial species that are rare and only occasionally found in clinical specimens, The majority of them are challenging to routinely identify, and some of them have poor documentation in a clinical sample. Also had an equal or greater impact on the extent of the disorder than usual species of bacteria [1]. The uncommon bacterial species infrequently exist in medical specimens such as Pseudomonas fluorescens [2] , Sphingomonas pauci-mobilis [3], Kocuria kristinae [4,5].
On the other side, the massive development and rise of multidrug-resistant (MDR) bacteria in numerous pathogenic bacteria, including such uncommon bacterial species are significant public health challenges [6].When an isolate was resistant to at least one agent in more than three antimicrobial groups or cat
Issue 30. February 2024 | Cardiometry | 41
egories, it was designated as MDR and if a bacterial isolate is still responsive to only one or two classes of antibiotics, it’s considered to be extensively drug-resistant (XDR). Isolate was designated pandrug-resistant (PDR) if it was resistant to all drugs in all antibacterial groups [7,8]. The critical degree of resistance contains Acinetobacter baumannii and pseudomonas aeruginosa which are resistant to carbapenems only. Thus, Klebsiella pneumonia, Escherichia coli, Entero-bacter spp., Serratia spp., Proteus spp. Providencia spp., and Morganella spp.,are resistant to carbapenems and third generation cephalosporins. While the extensive level of resistance contains part of the bacterial species of Enterococcus faecium , which is vancomycin-resistant and Salmonella spp. , fluoroquinolone-resistant [9]. According to several surveillance studies, the MDR is relatively high in uncommon species [1, 10-12]. Though, studies on the Middle East, particularly Iraq, are limited in focusing on the phenotype descriptions of some uncommon bacterial species. Hence, the current study aims to identify the prevalence of these species in clinical specimens and determine their profiles of antibiotic susceptibility.
MATERIALS AND METHODS
From the period June to October 2022, 52 uncommon bacterial isolates were identified in a variety of lesions including throat swab, sputum, blood, wound, abscess, stool, and urine from the patients admitted to Al Zahraa Teaching hospital for Maternity and Children in Najaf. Bacterial isolates were isolated according to the classical morphological (Gram stain/ Hime-dia, India) [13-15] and VITEK 2 system (bioMérieux, France) in addition to the measurement of Minimum Inhibitory Concentration (MIC). So profiles of antibiotic sensitivity of the bacterial isolates conducted detection to susceptibility examination by adjusted disc-diffusion method (Kirby-Bauer) [16-18] on Mueller- Hinton agar(MHA) (Oxoid, UK) plates were cultured by sterile swab plunged into the inoculums (0.5 McFarland).
The choice of antibiotic disc and isolates that were regarded resistant or susceptible were performed depending on [15,19-21] .The isolates susceptibility to the following antibiotics was examined: penicillin (P,1IU), Ampicillin (AMP,10μg), Piperacillin (PRL,100μg), Ticarcillin (TC,75μg), Ticarcillin/ clavulanic acid (TCC,85μg), Ampicillin/sulbactam (SAM,20μg), Piperacillin/tazobactam (PTZ,110μg), 42 | Cardiometry | Issue 30. February 2024
amoxicillin/clavulanic acid (AMC,30μg), Cefazolin (CZ,30μg ), Cephalexin (CL,30μg), Cefoxitin (FOX-,30μg), Cefpodoxime (CPD,30μg), Cefuroxime (CXM,30μg), Ceftazidime (CAZ,30μg), Cefotaxime (CTX,30μg), Ceftriaxone (CRO,30μg), Cefix-ime (CFM,5μg) , Cefepime (FEP,30μg), Aztreonam (ATM,30μg), Meropenem (MEM,10μg), Imipen-em (IPM,10μg), Ertapenem (ETP,10μg), Tobramycin (TOB,10μg), Amikacin (AK,30μg), Gentamicin (GM,10μg), Netilmicin (NET,30μg), Norfloxacin (NOR,10μg), Ofloxacin (OFX,5μg), Ciprofloxacin (CIP,5μg), Levofloxacin (LEV,5μg), Nalidixic acid (NA,30μg), moxifloxacin (MFX,5μg), Tetracycline (T,30μg), Minocycline(MI,30μg), Tigecycline (TG-C,15μg), Nitrofurantoin (NIT,300μg), Azithromycin (ATH,15μg), Trimethoprim (TM,5μg), Trimetho-prim/sulfamethoxazole (TS,25μg), Chloramphenicol (C,30μg), Colistin(CT,10μg), clindamycin (CD,2μg), linezolid(LNZ,10μg), vancomycin (VA,30μg), erythromycin (E,15μg), oxacillin (OX,5μg), teicoplanin TEC,30μg), rifampin (RA,5μg), fusidic acid (FA,10μg) (Himedia, India, Bioanlyse, Turkey and Mast Di-agnotics, UK). For objective of this study, the isolates were regarded a (MDR), (XDR), and (PDR) in Gram-negative isolates on the basis of standardized international criteria [22,23]. Finally, multiple antibiotic resistance (MAR) index of Seventeen gram-positive and gram-negative bacterial rare were detected by division of the number of medications against which strain demonstrated resistance of them above the total number of tested drugs [16]. Isolates showing intermediate susceptibility were considered as resistant. The control positive was applied E. coli ATCC 25922 [24].
RESULTS
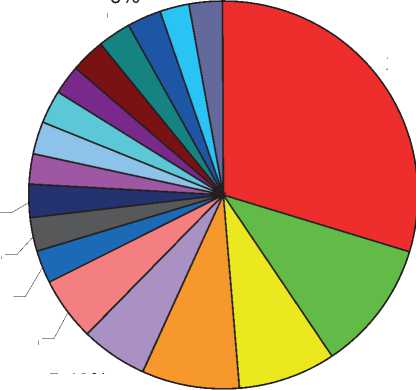
Fifty-two uncommon bacterial isolates acquired from patients who were complaining from different infections. Of these 37(71.2%) isolates found to be gram-negative bacteria and 15(28.8%) isolates found to be gram-positive bacteria. The current study showed that from the overall of 37 gram-negative bacteria, 11 (29.8%) isolates were identified as Acine-tobacter baumanii, 4(10.8%) Morganella morganii , 3(8.1%) Acinetobacter lwoffii , 3(8.1%) Providencia rettgeri , 2(5.4%) Salmonella typhi , 2(5.4%) Burkholde-ria cepacia group, 1(2.7%) as well as all of Salmonella paratyphi B, Salmonella enteritidis, Ochrabacturm an-thropi, Oligella ureolytica, Burkholderia mutivorans,
Serratia marcescens, Serratia fonticola, Sphingomonas paucimobilis, Pseudomonas fluorescens, Escherichia vulneris, Enterobacter cloacae complex, and Proteus penneri (Figure 1). While gram positive bacteria, 3(20%) isolates belong to Staphyllococcus haemolyt- icus, 3(20%) Streptococcus mutans, 3(20%) Kocuria kristinae, and 1(6.7%) for each Streptococcus uberis, Streptococcus mitis, Kocuria rhizophila, Kocuria rosea, Granuleticatella adiacens, and Leuconostoc mesen-teroides sub sp. cremoris (Figure 2).
-
□ A.baumannii
-
□ M. morganii
-
□ A. lwoffii
-
□ P. rettgeri
-
□ S. typhi
-
□ B. cepacia group
-
□ B. multivorans
-
□ S. enteritidis
-
□ S. paratyphi B
-
□ O. ureolytica
-
□ O. anthropi
-
□ S. marcescens
-
□ S. fonticola
-
■ S. paucimobilis
-
□ P. fluorescens
-
□ E. vulneris
-
□ E. cloacae complex
-
□ P. penneri
2.70%
2.70%
2.70%
2.70%
2.70%
2.70%
2.70%
2.70%
2.70%
29.80%
10.80%
5.40%
5.40%
8.10%
8.10%
Figure 1: The growth pattern of different members of uncommon gram-negative bacteria isolated from various lesions.
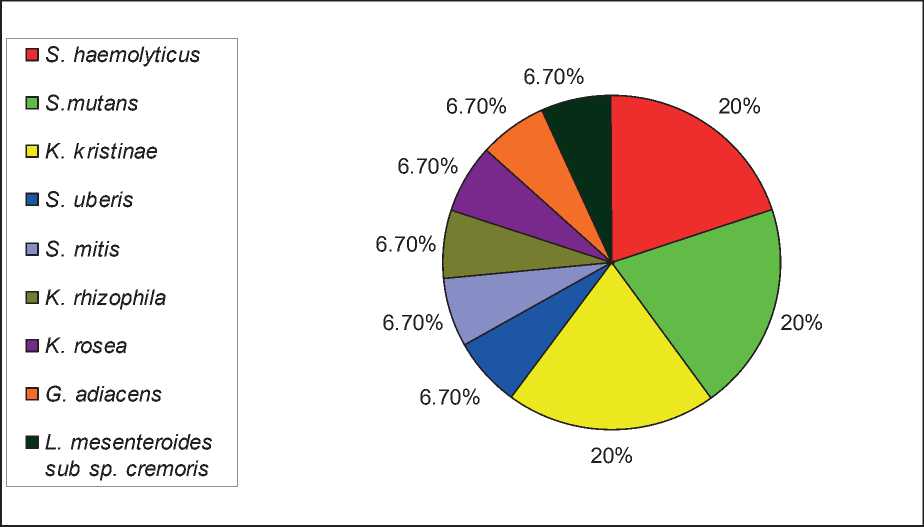
Figure 2: The growth pattern of different members of uncommon gram-positive bacteria isolated from various lesions.
All A. baumannii, M. morganii, P. rettgeri, and S. marcescens isolates were examined for their susceptibility of antibiotics against the selected 38 antibiotics by disc-diffusion method (Kirby-Bauer). Table (1)
gives resistance (R), the intermediate resistance (I), and the susceptibility (S) of the above isolated bacteria to various antibiotics as denoted by diameter in millimeters. The highlight marks cases considered
Table 1
Susceptibility patterns of some uncommon-gram-negative bacteria (UGNB)
|
Antibiotic |
Susceptibility profile % |
|||||||||
|
A.baumannii N(11) |
M. morganii N(4) |
P. rettgeri N(3) |
S. marcescens N(1) |
|||||||
|
R |
S |
R |
I |
S |
R |
I |
S |
R |
S |
|
|
Ampicillin |
100 |
- |
- |
100 |
- |
- |
100 |
- |
||
|
Piperacillin |
100 |
- |
100 |
- |
||||||
|
Ticarcillin |
100 |
- |
||||||||
|
Amoxicillin/clavulanic acid |
100 |
- |
- |
100 |
- |
- |
100 |
- |
||
|
Ticarcillin/ clavulanic acid |
100 |
- |
- |
50 |
50 |
- |
- |
100 |
100 |
- |
|
Ampicillin/sulbactam |
100 |
- |
50 |
25 |
25 |
- |
- |
100 |
100 |
- |
|
Piperacillin/tazobactam |
54.4 |
45.6 |
25 |
50 |
25 |
33.3 |
66.7 |
- |
100 |
- |
|
Cephalexin |
75 |
25 |
- |
66.7 |
33.3 |
100 |
- |
|||
|
Cefoxitin |
100 |
- |
50 |
50 |
- |
- |
- |
100 |
100 |
- |
|
Cefpodoxime |
100 |
- |
||||||||
|
Cefuroxime |
100 |
- |
- |
- |
- |
100 |
100 |
- |
||
|
Ceftazidime |
63.6 |
36.4 |
- |
25 |
75 |
- |
- |
100 |
100 |
|
|
Ceftriaxone |
100 |
- |
25 |
- |
27 |
- |
- |
100 |
100 |
|
|
Cefotaxime |
100 |
- |
25 |
- |
27 |
- |
- |
100 |
100 |
|
|
Cefixime |
25 |
- |
27 |
- |
- |
100 |
100 |
- |
||
|
Cefepime |
100 |
- |
- |
50 |
50 |
- |
- |
100 |
100 |
- |
|
Aztreonam |
- |
- |
100 |
- |
- |
100 |
100 |
- |
||
|
Imipenem |
36.4 |
63.6 |
25 |
50 |
25 |
- |
66.7 |
33.3 |
- |
100 |
|
Meropenem |
36.4 |
63.6 |
- |
- |
100 |
- |
- |
100 |
- |
100 |
|
Amikacin |
100 |
- |
- |
25 |
75 |
- |
- |
100 |
100 |
- |
|
Gentamicin |
44.4 |
55.6 |
- |
25 |
75 |
- |
- |
100 |
100 |
- |
|
Tobramycin |
44.4 |
55.6 |
25 |
- |
75 |
- |
- |
100 |
100 |
- |
|
Netilmicin |
36.4 |
63.6 |
100 |
- |
||||||
|
Nalidixic acid |
25 |
- |
75 |
- |
- |
100 |
100 |
- |
||
|
Ciprofloxacin |
54.4 |
45.6 |
25 |
- |
75 |
- |
- |
100 |
100 |
- |
|
Norfloxacin |
25 |
25 |
50 |
- |
- |
100 |
100 |
- |
||
|
Ofloxacin |
25 |
- |
75 |
- |
- |
100 |
100 |
- |
||
|
Levofloxacin |
36.4 |
63.6 |
25 |
- |
75 |
- |
- |
100 |
100 |
- |
|
Colistin |
63.6 |
36.4 |
||||||||
|
Polymyxin B |
77.8 |
22.2 |
||||||||
|
Tetracycline |
63.6 |
36.4 |
75 |
- |
25 |
100 |
- |
- |
100 |
- |
|
Doxycycline |
55.6 |
44.4 |
100 |
- |
||||||
|
Tigecycline |
- |
25 |
75 |
100 |
- |
- |
||||
|
Trimethoprim |
75 |
- |
25 |
- |
- |
100 |
100 |
- |
||
|
Trimethoprim/ Sulfamethoxazole |
44.4 |
55.6 |
50 |
25 |
25 |
- |
- |
100 |
100 |
- |
|
Chloramphenicol |
25 |
- |
75 |
- |
- |
100 |
100 |
- |
||
|
Nitrofurantoin |
75 |
- |
25 |
100 |
- |
- |
100 |
- |
||
|
Azithromycin |
100 |
- |
- |
100 |
- |
- |
100 |
- |
||
to be resistant to the specific antibiotics. The highest resistance (100%) of A. baumannii was to antibiotics (PRL, TCC, SAM, FOX, CRO, CTX, FEP, AK), and the lowest (36.4%) resistance to antibiotics (IPM, MEM, NET, LEV). Whereas, the highest resistance (100%) of M. morganii and P. rettgeri were to antibiotics (AMP, AMC, CXM, ATH) and ( AMP, AMC, T, TGC, NIT, ATH). Finally, S. marcescens was resistant (100%) to all antimicrobial agents excluding carbapenemes.
The MICs of all antibiotics for every bacterial isolate were determined by using VITEK2 system method. Depending on the results of MICs, the highest resistant UGNB are P. penneri followed S. fonticola then S. paratyphi B (Table 2).
The MICs of some uncommon-gram-positive bacteria (UGPB) showed that a first place in the resistance for S. haemolyticus followed by S. mitis and then S. mutans as shown in table 3.
Table 2
Minimum inhibitory concentration (MIC=μg/ml) dissemination of the UGNB (A: E. cloacae complex, B: P. penneri, C: P. fluorescens, D: E. vulneris, E: S. paratyhi B, F: S. enteritidis, G: S. typhi, H: S. paucimobilis, I: A. lwofii, J: S. fonticola, K: B. cepacia)
|
K |
J |
I |
H |
G |
F |
E |
D |
C |
B |
A |
Drugs |
|
- |
- |
- |
- |
- |
- |
- |
16 |
- |
- |
- |
AMP |
|
64 |
- |
- |
≥ 128 |
≤ 8-≥ 128 |
16 |
≤ 8 |
- |
≥ 128 |
≥ 128 |
- |
TC |
|
64 |
- |
- |
≤ 8 |
≤ 8 |
- |
≥ 128 |
≥ 128 |
- |
TCC |
||
|
64 |
- |
- |
≥ 128 |
≤ 4-≥ 128 |
≤ 4 |
≤ 4 |
- |
≥ 128 |
≥ 128 |
- |
PRL |
|
≥ 128 |
≤ 4 |
8, ≥ 128 |
≤ 4 |
≤ 4 |
≤ 4 |
≤ 4 |
≤ 4 |
≥ 128 |
≤ 4 |
≤ 4 |
PTZ |
|
- |
≥ 64 |
8, ≥ 64 |
- |
- |
- |
- |
≤ 4 |
- |
- |
≥ 64 |
CZ |
|
- |
8 |
- |
- |
- |
- |
- |
8 |
- |
- |
≥64 |
FOX |
|
4 |
32 |
2 |
32 |
≤ 1 |
≤ 1 |
≤ 1 |
≤ 1 |
32 |
≥ 64 |
≤ 1 |
CAZ |
|
- |
≥ 64 |
8 |
- |
- |
- |
- |
≤ 1 |
- |
- |
≤ 1 |
CRO |
|
4 |
4 |
≤ 1 |
4 |
≤ 1 |
≤ 1 |
≤ 1 |
≤ 1 |
4 |
≥ 64 |
≤ 1 |
FEP |
|
≥ 64 |
- |
- |
8 |
≤ 1 |
≤ 1 |
≤ 1 |
- |
- |
≥ 64 |
- |
ATM |
|
8 |
8 |
≤ 0.25 |
≤ 0.25 |
≤ 0.25 |
≤ 0.25 |
≤ 0.25 |
≤ 0.25 |
≥ 16 |
≥ 16 |
0.5 |
IPM |
|
4 |
- |
- |
≤ 0.25 |
≤ 0.25 |
≤ 0.25 |
≤ 0.25 |
- |
≥ 16 |
≥ 16 |
- |
MEM |
|
- |
≥ 8 |
- |
- |
- |
- |
- |
≤ 0.5 |
- |
- |
≤ 0.5 |
ETP |
|
4 |
≤ 2 |
≤ 2 |
4 |
≤ 2 |
≤ 2 |
≤ 2 |
≤ 2 |
≥ 64 |
≥ 64 |
≤ 2 |
AK |
|
4 |
≤ 1 |
≤ 1 |
≤ 1 |
≤ 1 |
≤ 1 |
≤ 1 |
≤ 1 |
2 |
≥ 16 |
≤ 1 |
GM |
|
2 |
- |
- |
≤ 1 |
≤ 1-2 |
≤ 1 |
≤ 1 |
- |
2 |
≥ 16 |
- |
TOB |
|
2 |
≥ 4 |
≥ 4 |
≥ 4 |
≤ 0.25-0.5 |
≤ 0.25 |
≥ 4 |
≤ 0.25 |
≥ 4 |
≥ 4 |
≤ 0.25 |
CIP |
|
- |
4 |
4 |
- |
- |
- |
- |
≤ 0.12 |
- |
- |
≤ 0.12 |
LEV |
|
4 |
- |
- |
4 |
≤ 1-8 |
4 |
8 |
- |
≥ 16 |
≥ 16 |
- |
MI |
|
- |
2 |
≤ 0.5 |
- |
- |
- |
- |
≤ 0.5 |
- |
- |
≤ 0.5 |
TGC |
|
- |
128 |
- |
- |
- |
- |
- |
64 |
- |
- |
128 |
NIT |
|
80 |
≥ 320 |
≤ 20 |
≥ 320 |
≤ 20-320 |
≤ 20 |
≥ 320 |
≤ 20 |
≥ 160 |
160 |
≤ 20 |
TS |
|
≥16 |
- |
- |
≤0.5 |
- |
- |
- |
- |
- |
- |
- |
CT |
Table 3
Minimum inhibitory concentration (MIC = μg/ml) dissemination of the UGPB (S. mitis, S. mutans, and S. haemolyticus).
|
S. haemolyticus |
S. mutans |
S. mitis |
Drugs |
|
≥ 0.5 |
0.25 |
≥ 8 |
P |
|
- |
≤ 0.25 |
8 |
AMP |
|
- |
≤ 0.12 |
≥ 8 |
CTX |
|
- |
0.25 |
4 |
CRO |
|
≤ 0.5-≥ 16 |
- |
- |
GM |
|
≤ 1-≥ 16 |
- |
- |
TOB |
|
≥ 4 |
- |
- |
OX |
|
0.25-≥ 8 |
≥ 16 |
0.5 |
LEV |
|
≤ 0.25-4 |
≥ 4 |
0.12 |
MXF |
|
≥ 8 |
≤ 0.12 |
- |
E |
|
≤ 0.25,≥ 8 |
≤ 0.25 |
0.5 |
CD |
|
,≥ 8 2 |
≤ 2 |
≤ 2 |
LNZ |
|
4, ≥ 32 |
- |
- |
TEC |
|
≥ 16 2- |
≥ 16 |
≥ 16 |
T |
|
≥ 32 -8 |
- |
- |
FA |
|
1-16 |
0.5 |
0.5 |
VA |
|
0.25-0.5 |
≤ 0.06 |
≤ 0.06 |
TGC |
|
- |
2 |
2 |
C |
|
≤16-256 |
- |
- |
NIT |
|
≤10- ≥ 320 |
- |
- |
TS |
|
≤0.5- ≥ 32 |
- |
- |
RA |
Among the A. baumannii isolates, 54.5%(6/11) were found to be MDR, and 18.2%(2/11) were XDR while 27.3%(3/11) were PDR. Also, P. penneri was found to be PDR, and S. marcescens and S. fonticola were XDR. Seventy five percent (3/4) of the M. mor-ganii isolates were MDR and the residual (25%) was XDR. One isolate of S. paratyphi B and S. typhi were MDR (Table 4, and 5).
All isolates were subjected to the calculation of the MAR index that showed high values greater than 0.2 rankings (0.21-0.94) from the highest MAR index (1) excluded E. vulneris (0.12) more details in table 6.
Table 4
Pandrug resistance (PDR), multi-drug resistance, and extensively drug-resistant (XDR) of some uncommon species.
|
Bacterial species |
No. (%) of MDR |
No. of bacterial isolates resistance to antibiotic classes |
No. (%) of XDR |
No. of resistance to antibiotic classes |
No. (%) of PDR-No. antibiotic classes |
|
S. paratyphi B |
1(100) |
1(3) |
- |
- |
- |
|
S. typhi |
1(50) |
1(4) |
- |
- |
- |
|
M. mor-ganii |
3(75) |
2(5)/1(3) |
1(25) |
8 |
- |
|
S. marc-escens |
- |
- |
1(100) |
9 |
- |
|
S. fonti-cola |
- |
- |
1(100) |
5 |
|
|
P. pen-neri |
- |
- |
- |
- |
1(100)-7 |
|
A. bau-mannii |
6(54.5) |
4(6), 2(5) |
2(18.2) |
2(8) |
3(27.3)- 9 |
Table 5
Multi-drug resistance and susceptibility of some uncommon bacterial species that were either not subject to or not present in Magiorakos et al.(2012)
|
Bacterial species |
No. of Multidrug resistance |
Types of antibiotic resistance |
Total drugs |
Types of antibiotic sensitive |
|
S. pauci-mobilis |
5 |
TC , PRL , CAZ , CIP , TS |
16 |
TCC , PTZ , FEP , ATM , IPM , MEM , AK , GM , TOB , MI , CT |
|
P. fluo-rescens |
11 |
TC , TCC , PRL , PTZ , CAZ , IPM , MEM , AK , CIP , MI , TS |
14 |
FEP , GM , TOB |
|
E. vulneris |
2 |
AMP , NIT |
16 |
PTZ , CZ , FOX , CAZ , CRO , FEP , IPM , ETP , AK , GM , CIP , LEV , TGC , TS |
|
E. cloacae complex |
3 |
CZ , FOX , NIT |
15 |
PTZ , CAZ , CRO , FEP , IPM , ETP , AK , GM , CIP , LEV , TGC , TS |
|
S. typhi |
3 |
AK , GM , TOB |
14 |
TC , PRL , PTZ , CAZ , FEP , ATM , IPM , MEM , CIP , MI , TS |
|
S. enterit-idis |
3 |
AK , GM , TOB |
14 |
TC , PRL , PTZ , CAZ , FEP , ATM , IPM , MEM , CIP , MI , TS |
|
Bacterial species |
No. of Multidrug resistance |
Types of antibiotic resistance |
Total drugs |
Types of antibiotic sensitive |
|
A. lwoffii |
4 |
PTZ , CZ , CIP , LEV |
12 |
CAZ , CRO , FEP , IPM , AK , GM , TGC , TS |
|
B. cepacia group |
CZ , CRO , FEP , GM , TS |
|||
|
B. cepacia |
13 |
TC , TCC , PRL , PTZ , FEP , ATM , IPM , AK , GM , TOB , CIP , TS , CT |
16 |
CAZ , MEM , MI |
|
B. mutiv-orans |
- |
- |
TS |
|
|
K. rhizo-phila |
- |
- |
RA , GM , CIP , PTZ |
|
|
S. uberis |
- |
- |
CRO , GM , CIP , PTZ |
|
|
S. mitis |
6 |
P , AMP , CTX , CRO , CD , T |
12 |
LEV , MXF-LNZ , VA , TGC , C |
|
S. mutans |
4 |
P , LEV , MXF , T |
13 |
AMP , CTX , CRO , E , CD , LNZ , VA , TGC , C |
|
S. haemo-lyticus |
11 |
P , OX , E , CD , TEC , VA , T , NIT , FA , RA , LNZ |
17 |
GM , TOB , LEV , MXF , TGC , TS |
|
S. haemo-lyticus |
9 |
P , OX , GM , TOB , LEV , MXF , E , FA , TS |
17 |
CD , LNZ , TEC , VA , T , TGC , NIT , RA |
|
S. haemo-lyticus |
8 |
P , OX , LEV , MXF , E , CD , T , FA |
17 |
GM , TOB , LNZ , TEC , VA , TGC , NIT , RA , TS |
(-)*: Missing
Table 6
MAR indices of all bacterial species
|
Bacterial species |
No. of Multidrug resistance |
Total drugs |
MAR indices |
|
M. morganii (4) |
11 , 12 , 14 , 22 |
29 |
0.83 , 0.41 , 0.48 , 0.76 |
|
P. rettgeri (3) |
7 , 8 , 8 |
29 |
0.24 , 0.27 , 0.27 |
|
S. marcescens |
32 |
35 |
0.91 |
|
S. paucimobilis |
5 |
16 |
0.31 |
|
P. fluorescens |
11 |
14 |
0.78 |
|
E. vulneris |
2 |
16 |
0.12 |
|
E. cloacae complex |
3 |
15 |
0.20 |
|
P. penneri |
14 |
15 |
0.94 |
|
Bacterial species |
No. of Multidrug resistance |
Total drugs |
MAR indices |
|
S. paratyphi B |
6 |
15 |
0.40 |
|
S. typhi |
3 , 7 |
14 |
0.21 |
|
S. enteritidis |
3 |
14 |
0.21 , 0.50 |
|
A. lwoffii |
4 |
12 |
0.34 |
|
S. fonticola |
9 |
15 |
0.60 |
|
B. cepacia group |
|||
|
B. cepacia |
13 |
16 |
0.81 |
|
B.multivorans |
|||
|
K. rhizophila |
|||
|
S. uberis |
|||
|
S. mitis |
6 |
12 |
0.50 |
|
S. mutans |
4 |
13 |
0.31 |
|
(3) S. haemolyticus |
8 , 9 , 11 |
17 |
0.47 , 0.52 , 0.65 |
DISCUSSION
The most common UGNB were Acinetobacter baumannii, Morganella morganii, Acinetobacter lwoffii, and Providencia rettgeri. UGPB were Staphylococcus haemolyticus, Streptococcus mutans, and Kocu-ria kristinae. In Iraq, many studies had documented the founding of multiple of these bacteria that have multiple drug resistance such as [10-12]. On the basis of standardized international criteria, the UGNB was classified into PDR, XDR, and MDR. Out of 20 UGNB belonging to seven species of UGNB, 11 were found to be MDR (55%), 5 XDR (25%), and 4 PDR (20%) (Table 4). The isolates were considered multidrug-resistant (MDR) if they were resistant to members of 3 or more of the above mentioned antibiotic classes. While an isolate that is resistant to all but one or two classes is defined as extensively drug-resistant (XDR), the definition of a pan-drug-resistant (PDR) in gram-negative strains is that isolates are resistant to all available classes of antibiotics [16]. A MAR index resistance to greater than 0.20 antibiotics reflects that bacteria arise where antibiotics are widely available in an environment, resulting in a high potential for wrong use and a ‘high-risk’ source of contamination ,There is insufficient data regarding the level of antibiotics resistance rare isolates associated with diverse infections in Iraq, hence may pose a public health provocation for clinicians. Thus, this study evaluated the MAR index of these isolates. However, the rising of MAR index values was noticed in our investigation ( Table 6), the vast majority of uncommon bacteria (96%, 24/25) had a MAR value of greater than 0.20, proving that there was widespread antibiotics usage and strong selection pressure in Najaf community. The MAR indices observed in this study are a likely sign that a significant proportion of the uncommon bacteria was present in response to many antimicrobial medications [16,17]. The high MAR obtained in the present study cautions us that any use of antibiotics in treatment should be preceded by an accurate identification of the underlying causes and an antimicrobial sensitivity test. This will not only help these medications be used more effectively, but it will also help reduce the number of antibiotic-resistant isolates in hospitals and communities in Iraq.
CONCLUSION:
The high MAR identified and found in PDR, XDR, and, MDR of uncommon bacteria in the current analysis makes it essential for antibiotic susceptibility testing to be done prior to antimicrobial treatment. This would not only assist in the appropriate use of antibiotics but also restrict the spread of antibiotic-resistant isolates in the hospitals of Iraq as well as in the community.
Limitations of the study
The first limitation of the study is the small sample size. We use a limited number to reduce the cost of the study due to the lack of a funder for the study. Also, it is recommended to make a follow-up study to add more information to the present case-control study.
Author’s contributions
All authors collaborated equally in all the research steps.
Human and animal rights
The research was carried out in compliance with the ethical and privacy regulations of Iraq and international standards, and adhered to the ethical principles outlined in the World Medical Association Declaration of Helsinki. In addition, it is noteworthy that our Institutional Review Board (IRB) adheres to the International Guideline for Human Research protection, which is mandated by the Declaration of Helsinki, The Belmont Report, the CIOMS Guideline, and 48 | Cardiometry | Issue 30. February 2024
the International Conference on Harmonization in Good Clinical Practice (ICH-GCP).
Consent for publication
All participants gave written informed consent before participating in this study.
Funding
There was no specific funding for this specific study.
Conflict of interest
The authors have no conflict of interest with any commercial or other association connected with the submitted article.
Acknowledgments
We thank the staff of Al-Zahraa Teaching Hospital for Maternity and Children in Najaf Governorate-Iraq for their help in collecting samples. We also thank the hospital’s internal labs’ high-skilled staff for their help in estimating biomarkers levels.
Список литературы Prevalence of pan drug-resistant, extensively drug-resistant, and detection of multiple antibiotic resistance in uncommon bacterial isolates in Al-Al Zahraa teaching hospital for maternity and children in Najaf/Iraq
- Faraj RK , Maarof MN. Isolation and identification of some uncommon bacterial species isolated from different clinical sample. .Journal of University Garmian, Conference Paper, 2017, 2:563-580. https://doi.org/10.24271/garmian.165.
- Morgan, RL Wang PW. The evolution of Pseudomonas syringae host specificity and type III effector repertoires. Mol Plant Pathol. 2009,10(6):307-320. 10.1111/j.1364-3703.2009.00587.x.
- Bhatia R, Tomar J. Sphingomonas paucimobilis – an emerging pathogen. IntJContempPediatr. 2016, 3(3):123-1125.http://dx.doi.org/10.18203/2349-3291.ijcp20162407.
- Tewari R, Dudeja M. Das AK, Nandy S. Kocuria kristinae in catheter associated urinary tract infection: a case report. Journal of Clinical and Diagnostic Research. 2013,7(8):1692-1693. 10.7860/JCDR/2013/6077.3247.
- Chen H, Chi H, Chiu N, Huang F. Kocuria kristinaeA true pathogen in pediatric patients. Journal of Microbiology, Immunology and Infection. 2015.48: 80-84. 10.1016/j.jmii.2013.07.001.
- Tshisevhe VS, Lekalakala MR, Tshuma,N, JanseVRS, Mbelle N. Outbreak of carbapenem-resistant Providencia rettgeri in a tertiary hospital. South Afr. Med. J.(SAMJ),2017,107(1):31-33. http://dx.doi.org/10.7196/samj.2017.v107i1.12002.
- Magiorakos AP, Srinivasan A.,Carey RB, Carmeli Y, Falagas ME, Giske CG. Multi drug resistant, extensively drug-resistant and pan drug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2022,18(3):268-281. https://doi.org/10.1111/j.1469-0691.2011.03570.x.
- Ammar Abdul Aziz Alibeg1*, Abbas H. Abdulsada2, Noor H. Nasser3, Karrar Abdil Aziz Alibeg. Design and Synthesis of Possible Mutual Prodrugs of (Nsaid) Etodolac and Tolmetin with (Cytotoxic) Gemcitabine Sys Rev Pharm 2020;11(11):315-318 A multifaceted review journal in the field of pharmacy.
- Soltani E, Hasani Am Rezaee MA, Pirzadeh T. Virulence characterization of Klebsiella pneumonia and its relation with ESBL and AmpC beta-lactamase associated resistance. Iranian Journal of Microbiology. 2020,12(2): 98–106. https://doi.org/10.18502/ijm.v12i2.2613.
- Arega B, Wolde-Amanuel Y, Adane K, Belay E, Abubeke A , Asrat, D. Rare bacterial isolates causing bloodstream infections in Ethiopian patients with cancer. Infect Agents Cancer. 2017,12:40. DOI: 10.1186/s13027-017-0150-9.
- Al-Mayahi FSA. ,Ali RH. Preliminary study of emergence MDR of pro videncia spp. isolates producing ESBL, AmpC and MBL among patients with RTI and in wastewater in AL-Diwaniya city, Iraq. Biochem. cell. arch.2018,18, Supplement 1: 1357-1368. doi: 10.3390/biology10111144.
- Al-Mayahi FSA, Jaber SM. A preliminary study of multiple antibiotic resistance (MAR) and extensively drug-resistant (XDR) of bacterial causing typhoid fever isolated from stool specimens in Al-Diwaniya, Iraq. Eurasia J. Biosci.2020a,14: 2369-2378. doi: 10.3390/tropicalmed7100271
- Al-Mayahi FSA, Jaber SM. Multiple drug resistance of Listeria monocytogenes isolated from aborted women by using serological and molecular techniques in Diwaniyah city/Iraq. Ran. Iran J. Microbiol. 2020,12(4):305-312. DOI: 10.18502/ijm.v12i4.3933.
- Forbes BA,Sahm DF,Weissfeld AS.Bailey and Scott’s Diagnostic Microbiology, Mosby Inc, Maryland Heights, Mo, USA,12th edition.2007,p1212-1214. DOI: 10.12691/ajidm-3-3-4.
- Oladeinde A , Abdo Z, Zwirzitz B, Woyda R, Lakin SM., Press MO.Litter Commensal Bacteria Can Limit the Horizontal Gene Transfer of Antimicrobial Resistance to Salmonella in Chickens. Applied and Environmental Microbiology.2022 ,88(9):76-83. doi: 10.1128/aem.02517-21.
- Liu X., Zhang J, Li Y, Shen Q, Jiang W, Zhao K. Diversity and frequency of resistance and virulence genes in blaKPC and blaNDM co-producing Klebsiella pneumonia strains from China. Infection Drug Resistance, 2019 12: 2819–2826. doi: 10.2147/IDR.S214960.
- Bauer AW, Kirby WMM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disc method. Amer. J. Clin. Pathol.2016,45:493-496. doi: 10.1128/aac.1.6.451
- Martin RM, Bachman MA Colonization, Infection, and the Accessory Genome of Klebsiella pneumoniae. Frontiers in Cellular and Infection Microbiology.2018:8:4. DOI: 10.3389/fcimb.2018.00004.
- Osama DM, Zaki BM, Khalaf WS. Mohamed MYA,Tawfick MM, Amin HA Occurrence and Molecular Study of Hypermucoviscous Hyper Virulence Trait in Gut Commensal K. pneumoniae from Healthy Subjects. Microorganisms, 2023,11(3): 704-710. https://doi.org/10.3390/microorganisms11030704.
- Asif M, Alvi I.A, Rehman SU. Insight into Acinetobacter baumannii: Pathogenesis, global resistance, mechanisms of resistance, treatment options, and alternative modalities. Infect.Drug Resist. 2018,11:1249-1254.doi: 10.2147/IDR.S166750.
- Nguyen M, Joshi SG Carbapenems resistance in Acinetobacter baumannii, and their importance in hospital‐acquired infections:a scientific review. Journal of Applied Microbiology. 2021, 131(6): 2715–2738. DOI: 10.1111/jam.15130.
- 10-Manenzhe RI, Zar HJ, Nicol MP, Kaba M. The spread of carbapenemase-producing bacteria in Africa A systematic review. J. Antimicrob. Chemother. 2019, 70: 23–40. DOI: 10.1093/jac/dku356.
- Al-Ghazaly NFZ. Molecular investigation of AmpC-Family and Sulfa Drugs Resistance Genes among Acinetobacter Spp. Isolated from Different Clinical Sources in Al–Najaf City.2022, p.129- 133. DOI: 10.53730/ijhs.v6nS1.7212.
- Russo, T.A.; Marr CM. Hyper virulent Klebsiella pneumonia. Clinical Microbiology Reviews. 2019, 32(3):33-39. DOI: 10.1128/CMR.00001-19.
- Howard A, Donoghue M, Feeney A, Sleator RD. Acinetobacter baumannii: An emerging opportunistic pathogen. Virulence 2012, 3: 243–250. DOI: 10.4161/viru.19700


