The paleogenetic study of Bertek-33, an Afanasyevo cemetery on the Ukok plateau, the Altai Mountains
Автор: Pilipenko A.S., Trapezov R.O., Cherdantsev S.V., Pilipenko I.V., Zhuravlev A.A., Pristyazhnyuk M.S., Molodin V.I.
Журнал: Archaeology, Ethnology & Anthropology of Eurasia @journal-aeae-en
Рубрика: Anthropology and paleogenetics
Статья в выпуске: 4 т.48, 2020 года.
Бесплатный доступ
We present the results of a paleogenetic analysis of bone samples representing seven adult individuals from Bertek-33—an Afanasyevo cemetery on the Ukok plateau, in the Altai Republic, Russia. The fi ndings are interpreted with reference to archaeological and anthropological data. Four systems of genetic markers were analyzed: mitochondrial DNA, polymorphic fragment of the amelogenin gene, autosomal STR-loci, and Y-chromosomal STR-loci. Genetic results indicate the dominance of Western Eurasian mtDNA haplogroups (T, J, U5a, K, H) and the homogeneity of the male gene-pool represented by variants of the Y-chromosomal haplogroup R1b. Data on mtDNA, Y-chromosome, and individual autosomal markers attest to the Western Eurasian affi nities of this group. The sample falls within the mtDNA and Y-chromosomal diversity of the Afanasyevo population of southern Siberia. Possible kinship between the individuals buried at Bertek-33 is discussed. Also, we address theoretical issues such as the accuracy of comparisons and the interpretation of genetic data with regard to cultural features.
Paleogenetics, Afanasyevo culture, mitochondrial DNA, Y-chromosome, Altai Mountains, Bronze Age
Короткий адрес: https://sciup.org/145146037
IDR: 145146037 | DOI: 10.17746/1563-0110.2020.48.4.146-154
Текст статьи The paleogenetic study of Bertek-33, an Afanasyevo cemetery on the Ukok plateau, the Altai Mountains
The cemetery of Bertek-33 is situated on the left-bank side of the Bertek valley, at the first terrace of the Ak-Alakha River, at the Ukok plateau (the Altai Mountains, Russia) (Fig. 1). Five kurgans were studied at the site by the Western Siberian unit of the North Asian Joint Expedition of the IAET SB RAS. Four of these kurgans
had been excavated in 1991 under supervision of D.G. Savinov (1994a, b), while the fifth was studied later by V.I. Molodin. Kurgans 1–3 formed a compact chain, joining one another, and thus were excavated as a single unit (Fig. 2). Kurgan 4 was situated just a few meters from the first three mounds, forming a part of the same chain, while kurgan 5 was also very close to the others. Therefore, all the five objects were further treated as
Fig. 1. Location of the Bertek-33 site.
one burial site, Bertek-33. The grave-goods and burial customs observed in kurgans 1–4 were clearly typical of the Afanasyevo culture (Ibid.). The cultural affiliation of kurgan 5 could not be determined from the same indicators because of the substantial destruction of the complex by the water from the river, but it was tentatively assigned to the same archaeological culture. Thus, the skeletal individuals from kurgans 1–5 of Bertek-33 are considered as a single sample of the people of the Afanasyevo culture from southern Altai (Chikisheva, 1994, 2012: 66).
The remains of 8 adult individuals and an infant (fragmentary) were found in the kurgans. Kurgans 1, 3, and 4 yielded single burials; kurgan 2 contained burials of two adults and an infant; and kurgan 5, a collective burial of three adult individuals (Fig. 3, 4).
This paper outlines the results of a molecular genetic study of seven adult individuals from kurgans 2–5 at Bertek-33 (Table 1). On the basis
Fig. 2. Bertek-33 in the process of excavation.
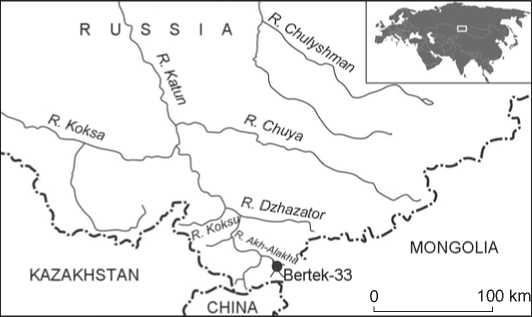

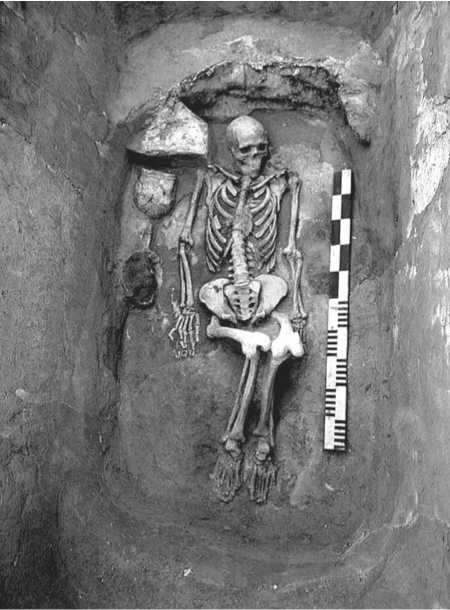
Fig. 3. Burial in kurgan 3.
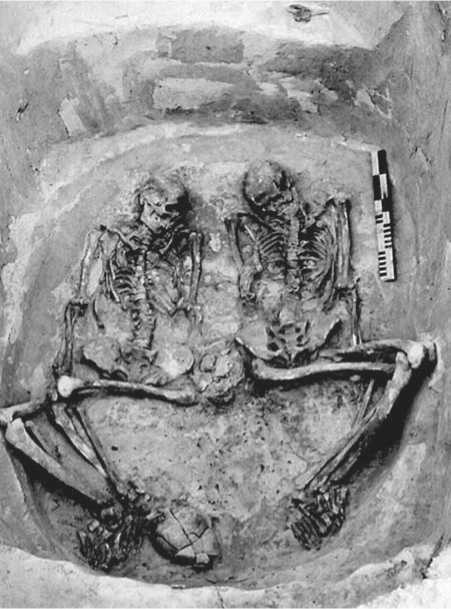
Fig. 4. Collective burial in kurgan 2.
Table 1. Description of the skeletal sample and the results of the analysis of the mtDNA structure
|
Number of individual |
Location |
Age at death* |
Haplotype of the mtDNA HVR I region |
Haplogroup (subgroup) of mtDNA |
|
1 |
Kurgan 2, burial 1, skeleton 1 |
20–25 |
16126C-16294T-16296T |
T |
|
2 |
Ditto, skeleton 2 |
25–28 |
16069T-16126C-16145A-16172C-16222T-16261T |
J (J1b1a1) |
|
3 |
Kurgan 3, burial 1 |
25–30 |
16224C-16291T-16311C-16362C |
K |
|
4 |
Kurgan 4, burial 1 |
Senilis |
16362C |
H |
|
5 |
Kurgan 5, burial 1, skeleton 1 |
25–30 |
16256T-16270T |
U5a |
|
6 |
Ditto, skeleton 2 |
40–45 |
16256T-16270T |
U5a |
|
7 |
Ditto, skeleton 3 |
50–60 |
16126C-16163G-16186T-16189C-16294T |
T1 |
*After (Chikisheva, 2012: 209, 213).
of the results obtained, a comparative analysis involving data on other local groups of the Afanasyevo people was carried out. We tested the correctness of assigning kurgan 5 to the Afanasyevo culture, and explored the kinship structure of the sample. It is of note that Bertek-33 is a completely excavated and studied archaeological complex representing a local Afanasyevo population, which was situated between groups of the same culture from the central part of the Altai Mountains (Vadetskaya, Polyakov, Stepanova, 2014) and its southern areas in North-Western Mongolia (Kovalev, Erdenebaatar, 2009).
Materials and methods
Samples for the molecular study were taken from the best-preserved (judging by macroscopic appearance) postcranial elements and teeth of the seven adult individuals from kurgans 2–5.
Preliminary treatment of the skeletal samples and DNA extraction. The methods applied in our previous publications were employed (Pilipenko, Trapezov, Zhuravlev et al., 2015; Pilipenko, Trapezov, Cherdantsev et al., 2018). In order to eliminate possible modern DNA contamination, the external surfaces of the samples were treated with 5 % sodium hypochlorite, and then irradiated with UV. The external bone layer (ca 1–2 mm thick) was mechanically removed, and then the sample was once again treated with UV. Fine bone powder was then drilled out from the cortical layer. The teeth were treated with 5 % sodium hypochlorite, mechanically cleared of external contaminants, irradiated with UV, and ground down using the vibration orbicular grinder Retsch MM200 (Germany).
Before DNA extraction, the bone powder (postcranial samples) was incubated in a 5M guanidine thiocyanate buffer at 65 ºC and constantly mixed during incubation. The teeth specimens were decalcified with 0.5M EDTA solution, followed by lysis with proteinase K. DNA extraction was performed employing a phenol/chloroform protocol, with subsequent sedimentation with isopropanol.
Analysis of genetic markers. Four systems of molecular genetic markers were analyzed: mtDNA (HVR I region), a fragment of an amelogenin gene (sex marker), highly variable autosomal STR-loci (universal markers of the degree of kinship), and STR-loci of the Y-chromosome— phylogenetically and phylogeographically informative markers of the male line of kinship (Pilipenko et al., 2017).
Amplification of the mtDNA HVR I region was performed using two different protocols: four short overlapping fragments using one-cycle PCR (Haak et al., 2005) and one long fragment using two-cycle nested PCR (Pilipenko et al., 2008). DNA sequencing was carried out with an ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, USA). Sequencing extracts were analyzed with an ABI Prism 3100XL Genetic Analyzer automatic capillary sequencer (Applied Biosystems, USA) at the SB RAS Genomics Core Facility . The obtained results were interpreted using phylogenetic and phylogeographic analysis, as described earlier (Pilipenko, Trapezov, Polosmak, 2015).
Profiling of 15 autosomal STR-loci and analysis of the amelogenin gene region polymorphism was performed using the AmpFlSTR® Profiler® Plus PCR Amplification Kit (Applied Biosystems, USA), following the manufacturer’s protocol. Profiles of 17 STR-loci of the Y-chromosome were determined using the commercial AmpFlSTR® Y-filer® PCR Amplification Kit (Applied Biosystems, USA), also following the manufacturer’s protocol. Haplogroups of the STR-haplotypes of the Y-chromosome were determined using two freeware programs: Haplogroup predictor hapest5/) and Vadim Yurasin’s YPredictor 1.5.0 .
Anti-contamination measures and verification of the results. All procedures with the skeletal specimens were carried out in a specially-equipped laboratory for molecular paleogenetics (Institute of Cytology and Genetics of the SB RAS, Novosibirsk, Russia). A description of the anti-contamination measures and verification of the results can be found in our previous publication (Pilipenko et al., 2018).
Results and discussion
Degree of DNA preservation. The climatic conditions of the Altai Mountains, including the Ukok plateau, are favorable for the preservation of ancient DNA in biological specimens from the archaeological sites of various historical periods (Pilipenko, Trapezov, Polosmak, 2015; Pilipenko et al., 2016). Our analysis has shown that the degree of DNA preservation in all the samples is predictably good; thus, a complete study of the structure of mtDNA could be performed. We successfully amplified mtDNA fragments of various lengths: from less than 150 to more than 300 basepairs. The study of allele profiles of the Y-chromosome STR- and autosomal loci, though more sensitive to the degree of ancient DNA’s preservation, was also possible and helped to reconstruct the genetic affinities of the individuals. These markers were best preserved for skeletons 3 and 4 (single burials in kurgans 3 and 4), where complete allele profiles of the 17 STR-loci of the Y-chromosome were obtained. Samples from the same individuals also produced almost complete allele profiles for the autosomal STR-loci: for 14 out of 15, excluding one locus with the longest PCR extract. The skeletons from a double burial in kurgan 2 (skeletons 1 and 2) and a collective burial in kurgan 3 (skeletons 5–7) displayed worse preservation of nuclear DNA, as was clear from the results of the analysis of the autosomal STR-loci allele profile: for only 8–12 out of 15 loci could their status be determined. It is of note that the reaction kit used for determining the Y-chromosome STR-loci profiles appeared to be less sensitive to the degree of DNA preservation as compared to the kit employed for the autosomal markers. Obviously, profiling of autosomal STR-loci alleles is the most objective indicator of the degree of nuclear DNA’s preservation in skeletal remains.
Substantial variation in the degree of DNA preservation in skeletons from different complexes of the same site obstructs employing complete archaeological samples for molecular genetic analyses, which decreases the value of such analyses. This variation may occur for several reasons: degradation of remains before inhumation, differences in the construction of burial complexes across the site, and various effects from destructive environmental factors. In the case of Bertek-33, the relatively poor DNA preservation in all the skeletons from kurgan 5 was not unexpected by us, since by the time of excavation the kurgan had been severely damaged owing to repeated destruction of the mound by the river. Infiltration of water has led to the degradation of the remains and, consequently, of the ancient DNA they contained. Thus, only the teeth from the individuals from kurgan 5 were employed in the study, as the bone elements most resistant to the influence of environmental destruction.
Importantly, all the specimens demonstrated the features typical of ancient DNA: better preservation of mtDNA as compared to nuclear markers, and an inverse correlation between the efficiency of amplification and the length of DNA fragments. This additionally verifies the correctness of the results obtained.
Sex determination and verification of the results. Determination of the sex of skeletal individuals by molecular genetic methods is a necessary part of a genetic study irrespective of the presence (or absence) of determination done using macroscopic methods, since this latter approach often produces incorrect results (Sierp, Henneberg, 2015; Gonzalez et al., 2017). Our results have shown that six out of the seven individuals from Bertek-33 were male (Table 2).
The high reliability of the molecular genetic data obtained was confirmed on the basis of the following criteria: concordance between the results of independent analyses of polymorphism of the amelogenin gene and allele profile of the Y-chromosome (both analyses of the presence/absence of Y-chromosome markers were performed for all the skeletons); presence of several structural variants of the Y-chromosome that belong to the same phylogenetic cluster; uniqueness (at the scale of the sample) of the autosomal allele profiles of the individuals; great diversity of mtDNA variants; identity of the results with multiple repetitions of the analysis (for each of the skeletons the analysis was carried out using four DNA extracts obtained at different times during about a two year period); absence of overlap between the structure of the genetic markers of the individuals buried at Bertek-33 and employees of the paleogenetic lab; and the presence of specific features typical of degraded ancient DNA from skeletal remains (see above).
Diversity of individual variants of the Y-chromosome and mtDNA. Reliable data on the structure of mtDNA were obtained for all seven skeletal individuals: the mtDNA HVR I region was sequenced, haplotypes reconstructed, and the phylogenetic position of the variants determined (Table 2). In total, six structural variants of mtDNA were identified, while identical variants were only found in skeletons 5 and 6 from the collective burial in kurgan 5. The variants belong to five haplogroups of mtDNA: T (two variants belonging to different subgroups),
Table 2. Results of the analysis of autosomal STR-loci, polymorphic fragment of the amelogenin gene and genetically determined sex of the deceased
|
X (D GO |
0) as e e e |
ф го Е 0) |
0) ГО е |
||||
|
и|иа6о|ашу |
X X |
||||||
|
VOd |
Q Z |
co CM co CM |
CM co |
см со см |
Q Z |
||
|
8L8SSO |
co cm |
CM О |
CM |
CM |
см |
& С) |
о С) |
|
isseia |
Q Z |
J |
co |
Q Z |
|||
|
XOdl |
Q Z |
CM 5) |
a> co |
to со |
го со |
Q Z |
|
|
VMA |
co LO |
LO LO |
co co |
co LO |
со |
со |
LO |
|
eevseia |
co co |
LO LO |
co |
О) |
со |
LO СО |
|
|
eeeLSza |
Q z |
= |
5= |
5 |
Q Z |
||
|
6ESS9LO |
Q Z |
CM & |
co O) |
to |
о |
со о |
Q Z |
|
z i-esei-a |
to to |
co |
CM co |
см со |
ю см |
to со |
|
|
ЮН1 |
Q Z |
co ai co ci |
& co |
со ci со ci |
со ci со |
со ci со |
|
|
8881. sea |
co LO |
& LO |
co LO |
LO |
LO |
со со |
|
|
OdLdSO |
Q Z |
^ |
co CM |
Q z | |
Q Z |
||
|
ozesza |
о to |
о ai |
о co |
со |
со |
^ |
|
|
llslzq |
о co cm |
co CM co CM |
co co CM |
о co о co |
о со со см |
см со со со см |
со со |
|
6z и sea |
о о |
CM CM |
о |
о о |
см £? 52 |
см |
со |
|
S 3 1 1 |
^- |
CM |
co |
■* |
LO |
со |
1^ |
|
VHViVOA |
CO CM CO CO CO CO |
|
segsxa |
со со со co co 5 CM CM CM CM CM 2 |
|
ssvsao |
co co co co co co |
|
gsvsAa |
co co co co co co |
|
8»5А0 |
о о о о о Q |
|
eevsAa |
со со со со со 9 |
|
eevsAa |
9 СМ СМ СМ СМ 9 Z Z |
|
zevsAa |
LO LO LO LO LO LO |
|
eeesAa |
см см см см см см |
|
zeesAa |
СО СО СО СО 9 8 |
|
LeesAa |
^ ^ ^ ^ ^ ^: |
|
oeesAa |
со ю м- со м- 9 СМ СМ СМ СМ СМ 2 |
|
neeesAa |
О О О О О 9 СМ СМ СМ СМ СМ 2 |
|
leeesAa |
со со со со со со |
|
q/BseesAa |
|
|
6L SAO |
чг чг Ч|- Ч|- Ч|- ЧТ |
|
S 1 E о ^ 1 |
-^ СМ СО ЧГ СО h- |
J (one variant from the J1b1a1 subgroup), K and H (one variant of each), and U5a (one structural variant in two individuals). Thus, the phylogenetic diversity of mtDNA in the studied sample is substantial. An important result is that all the identified variants belong to the Western Eurasian cluster of mtDNA haplogroups.
Allele profiles of the Y-chromosome STR-loci were determined for the six male (based on molecular genetic data) individuals (Table 3). The number of STRloci for which reliable data could be obtained varied: a full profile was reconstructed for three individuals (2–4), while for two (1 and 6) only alleles of 16 out of 17 analyzed loci could be determined. For skeleton 7, which yielded poorly preserved DNA, only 10 loci were identified. Despite the difference in the completeness of the allele profiles of the STR-loci, the phylogenetic affiliations of all the Y-chromosome variants could be determined with high probability (from 99.3 to 100 %), using the predictor-software. Unlike the mtDNA sample collection discussed above, the Y-chromosome sample is fairly uniform: all the variants belong to the same phylogenetic cluster—the R1b haplogroup. Moreover, the status of only two loci of the Y-chromosome among all the allele profiles was found to be variable: DYS390 and YGATAH4. The remaining 15 exhibit identical alleles in all the studied specimens (with allowance for the absence of data on some loci for some of the specimens). In five specimens with complete (or almost complete) allele profiles, not less than three different structural variants of the Y-chromosome belonging to the R1b haplogroup were identified: a unique (for this particular sample) variant in skeleton 2, and two variants in pairs of skeletons—1 and 4, 3 and 6.
Clearly, such a small group of individuals cannot be considered representative of the contemporaneous Afanasyevo population of the Altai Mountains (or even of part of it). Nevertheless, some common features are worth pointing out. First, the domination of Western Eurasian variants (though also variable) of mtDNA haplogroups. Second, the phylogenetic and, to a substantial degree, structural uniformity of variants of the Y-chromosome. No “genetically contrast”, or outlying, individuals were identified.
Notably, no burial complexes from the Afanasyevo culture have yet been detected in the territory of the Altai Mountains and Mongolia neighboring the Ukok plateau. This fact emphasizes the value of the paleogenetic data obtained in our study for exploring the variation of the genetic structure of local groups from this culture. A comparative analysis involving the samples of the mtDNA and Y-chromosome specimens from Bertek-33 and other Afanasyevo sites from the Altai Mountains and Minusinsk Basin has been carried out. The analysis includes both previously published data (Allentoft et al., 2015; Hollard et al., 2018) and our unpublished results. This analysis has shown the overwhelming prevalence of Western Eurasian mtDNA haplogroups in all the population groups. Moreover, all the mtDNA haplogroups (and even identical mtDNA variants in some cases) detected in the sample from Bertek-33 had been previously found in specimens from other Afanasyevo sites in the Altai Mountains and Minusinsk Basin. It is of note, however, that the mtDNA variants identified in the specimens from Bertek-33, taken separately, are not specific to the Afanasyevo population only, but rather, quite widespread across Western Eurasia.
The similarity between the sample from Bertek-33 and other Afanasyevo sites is even stronger in terms of the composition of variants of the Y-chromosome: almost all of them belong to the R1b haplogroup. Its predominance is a unique feature of the Afanasyevo population of the region, which distinguishes it from other southern Siberian Bronze Age populations, as well as from later groups. Among contemporaneous Bronze Age populations outside southern Siberia whose Y-chromosome gene-pool has been studied to date, such a feature was observed in some local groups from the Yamnaya culture in Eastern Europe (Haak et al., 2015; Allentoft et al., 2015).
The similarity between the individuals from Bertek-33 and other local groups from the Afanasyevo culture is paralleled by observations made by physical anthropologists. According to the results of a craniometric study, the skulls from this burial site display similarity to the Afanasyevo cranial samples from the highlands of the southwestern and central areas of the Altai Mountains, and also to the easternmost Afanasyevo sample from Xinjiang (Chikisheva, 1994: 166). As regards the Y-chromosome and mtDNA data, we did not detect any genetic trace of admixture with the autochthonous population of this region. By “autochthonous” we mean the pre-Afanasyevo populations of Altai and the neighboring areas of southern Siberia and Central Asia, not connected genetically with the western part of Eurasia. Such an influence of the autochthonous groups, connected to the so-called southern Eurasian anthropological formation, on the Afanasyevo population from Bertek-33 had been previously detected in craniometric data (Chikisheva, 2012: 67). Notably, the most evident manifestations of this admixture were observed in the adult individual from kurgan 1, who was not analyzed in the present study.
There are other confirmations of the Western Eurasian vector of genetic connections of the Bertek-33 population, apart from the Y-chromosome and mtDNA data. A high frequency (more than 50 %) of the allele variant 9.3 of the THO1 STR-locus of the tyrosine hydroxylase 1 gene was observed in the sample. Such frequencies of this marker are typical of modern populations of the western part of Eurasia (Europe), while they are much rarer outside Europe (Brinkmann et al., 1996). Interestingly, this allele variant is considered by some scholars to be associated with longevity in European populations (Tan et al., 2002; Wurmb-Schwark et al., 2011). The high frequency of this marker is obviously one more independent piece of evidence supporting the Western Eurasian origin of the studied ancient population.
Thus, the studied sample from the Bertek-33 cemetery fits well into the range of intra-population mtDNA and Y-chromosome genetic variation of the Afanasyevo people in southern Siberia in general, which is in accordance with the findings of physical anthropology and archaeology (Chikisheva, 2012: 66; Molodin, 2001). Kurgan 5 deserves special attention, since owing to the great destruction of this complex it was not possible to describe the details of burial rites and grave goods. Therefore, this kurgan was only tentatively assigned to the Afanasyevo culture. The results of the present study confirm such an attribution of the complex. It is important to keep in mind, however, that genetic markers by themselves cannot be used for confirmation of the cultural affinities of burial complexes, since they are nothing more than specific features of a person as a biological individual. These individual markers can be considered in the light of their compliance (or non-compliance) with the intragroup genetic variation of the ancient population to which the individual potentially belonged. If some specific genetic features were established for a population, the presence of these features in an individual (or individuals) might serve as an important, but inconclusive, argument supporting his/her/their assignment to this particular population. It should be kept in mind that the terms “biological population” and “people of an archaeological culture” are never identical. The presence of carriers of various cultural traditions in a biologically uniform population is quite a typical situation; and vice-versa, a population homogeneous in terms of material culture might well include a number of genetically diverse groups.
Only in some cases can genetic data be used for indirect (!) evaluation of the correctness of a cultural attribution of an archaeological complex. For example, the presence of the Y-chromosome and mtDNA variants typical of the Afanasyevo population of southern Siberia in the skeletons from kurgan 5 at Bertek-33, together with the position of the complex in the chain formed by kurgans 1–4, are an additional indirect evidence for considering all the five kurgans as one cemetery belonging to the Afanasyevo culture. This conclusion is also confirmed by the results of the craniometric study showing that all the skeletal individuals form a single sample (Chikisheva, 2012: 66–67). An important argument in this discussion could be close kinship affiliations between individuals from various complexes of the same site. The presence of identical structural variants of the Y-chromosome and mtDNA can confirm direct kinship between individuals, as well as different types of close relatedness via paternal or maternal lines (respectively).
There are no data available about direct or close maternal kinship between the three individuals from kurgan 5 and other kurgans at Bertek-33. In the whole sample, the only case of a shared mtDNA variant (belonging to the U5a haplogroup) was detected in two individuals from the triple burial (skeletons 5 and 6). But the age at death of these individuals (25– 30 year-old female and 40–45year-old male) excludes the possibility of direct “mother-son” relatedness. However, another types of maternal kinship, e.g. “brothersister”, seem more plausible. A large proportion of allele variants of the autosomal STR-loci common between these two individuals additionally supports their kin relationship.
Let us turn to the structure of the Y-chromosome specimens of the five male skeletons from Bertek-33 from the point of view of paternal kinship. As was noted above, three different structural variants of the same R1b haplogroup were detected. Two pairs of the individuals display identical variants: skeletons 1 (kurgan 2) and 4 (kurgan 4); skeletons 3 (kurgan 3) and 6 (kurgan 5). This is an important piece of evidence supporting their patrilineality. In the case of the first pair (1 and 4), the probability of a direct “father-son” relation is fairly high: all the eight successfully genotyped autosomal STR-loci of these two individuals contain at least one common allele variant. The robustness of this conclusion is, however, weakened by the absence of data about the remaining seven loci that were not genotyped because of the poor preservation of the DNA in skeleton 1. The possible patrilineality of the second pair of individuals (3 and 6), exhibiting identical structural variants of the Y-chromosome, was clearly not a direct “father-son” kinship, since a number of the STR-loci display no common allele variant. Neither could they be full siblings, though other types of paternal kinship cannot be excluded, as is suggested by the presence of common alleles for many loci. Thus, both matrilineal and (likely more often) patrilineal kinship was an important factor determining the inhumation of Afanasyevo people at the same cemetery. However, this kinship might be not necessarily direct.
Conclusions
Our molecular genetic study of the individuals from Bertek-33 made a substantial contribution to the existing database of the Y-chromosome and mtDNA gene-pools of the Afanasyevo population of southern Siberia. Previously, genetic data had only been obtained for the population of the central part of the Altai Mountains and Minusinsk Basin. The results of the genetic study were thoroughly explored at different levels: single individual, burial complex, cemetery as a whole, and Afanasyevo populations of various scales. The interpretation of these data in the light of the results of archaeological and anthropological research has helped to resolve partially the long-standing questions of the genetic origin and connections of the Afanasyevo population of the Altai Mountains, its burial rites, and kinship structure. The composition of mtDNA and, particularly, Y-chromosome variants in the skeletal sample from Bertek-33 links this group to the bearers of the Afanasyevo culture from other regions of southern Siberia, and points towards their Western Eurasian origin. The genetic data suggest that kinship was an important factor in determining the inhumation of individuals in the same burial. Further progress in this area of research will require a substantial increase in the representativeness of the genetic databases describing local groups from the Afanasyevo culture, including high coverage whole-genome data for both single individuals and skeletal samples.
Acknowledgements
This study was supported by the Russian Science Foundation (Project No. 17-78-20193). The paleogenetic facilities of the Institute of Cytology and Genetics of the SB RAS are partially supported by Budget Project No. 0259-2019-0010-C-01.


