WEBspike: A New Proposition of Deterministic Finite Automata and Parallel Algorithm Based Web Application for EEG Spike Recognition
Автор: Anup Kumar Keshri, Shamba Chatterjee, Barda Nand Das, Rakesh Kumar Sinha
Журнал: International Journal of Information Technology and Computer Science(IJITCS) @ijitcs
Статья в выпуске: 11 Vol. 5, 2013 года.
Бесплатный доступ
The brain signal or Electroencephalogram (EEG) has been proved as one of the most important bio-signal that deals with a number of problems and disorders related to the human being. Epilepsy is one of the most commonly known disorders found in humans. The application of EEG in epilepsy related research and treatment is now a very common practice. Variety of smart tools and algorithms exist to assist the experts in taking decision related to the treatment to be provided to an epileptic patient. However, web based applications or tools are still needed that can assist those doctors and experts, who are not having such existing smart tools for EEG analysis with them. In the current work, a web based system named WEBspike has been proposed that breaks the geographical boundary in assisting doctors in taking proper and fast decision regarding the treatment of epileptic patient. The proposed system receives the EEG data from various users through internet and processes it for Epileptic Spike (ES) patterns present in it. It sends back a report to the user regarding the appearance of ES pattern present in the submitted EEG data. The average spike recognition rate obtained by the system with the test files, was 99.09% on an average.
Deterministic Finite Automata, Electroencephalogram, Epileptic Spike, Message Passing Interface, Parallel Computing, Scalability, World Wide Web
Короткий адрес: https://sciup.org/15011995
IDR: 15011995
Текст научной статьи WEBspike: A New Proposition of Deterministic Finite Automata and Parallel Algorithm Based Web Application for EEG Spike Recognition
Published Online October 2013 in MECS
Epilepsy is one of the most common brain disorder reported in human beings. It is not only very dangerous but may also lead to even death, if the patient is not treated in time. Therefore, early information about the occurrence of forthcoming Epileptic seizures becomes very important. Occurrences of sharp rise and fall patterns called Epileptic Spikes (ES) in the brain signals or Electroencephalogram (EEG) is one of the main symptom of epilepsy. By identifying or detecting the presence of ES patterns in EEG, an expert can take proper decision for treatment of an epileptic patient. Unfortunately, due to high sampling rate and longer duration of recording, the EEG recording produces enormous data [1]. Therefore, manual inspection of such massive data for specific pattern is not possible in real time.
Since its evolution, many automated tools and algorithms have been proposed by many researchers for automated recognition of ES patterns in EEG. These classifiers are based on wavelet transform [2-8], artificial neural networks [9-11], fuzzy logic [12] and non-conventional tools like finite automata [13, 14]. These methods are conventionally sequential in nature and hence, processing of massive data of recorded EEG becomes difficult. Parallel computing is other paradigm of computing where simultaneously multiple jobs are performed to save time or to do more work in less time. It can be applied to do a task that involves huge data processing. In the field of Biomedical Engineering, parallel computing can be applied for processing of various bio-signals, which are naturally huge in size. The authors have already demonstrated one such successful attempt earlier [15].
Availability of automated diagnostic tools with every doctor and the clinician is not assured. The doctors working in remote area may not have such tool with them. Therefore, to analyze the EEG of a patient, they may need to travel to some specialized diagnostic centres that further, may again increase the time of diagnosis. This delay in the treatment procedure may cause even death of the patient. Therefore, a smart system is needed, which can help doctors working in remote areas without having automated tools to analyze EEG. However, the review suggests that only few work in this aspect have been reported [16].
In the current work, a web-based application named WEBspike has been proposed that facilitates doctors working in different locations over the globe to submit the EEG of the patient to a centralised server through internet. The algorithm running in the server takes the submitted EEG data by the doctors, analyze them, and produces report that contains information about the occurrence of ES patterns in the EEG. These reports are then sent back to the doctors via e-mail, which ultimately help them in taking suitable action regarding the treatment of the epileptic patient. Multiple numbers of users can log to WEBspike simultaneously at a time and can submit the EEG data. Therefore, the volume of the submitted EEG data increases. To process such bulky EEG data in real time, parallel computing has been incorporated in the design. The server is connected with four other computers forming a multicomputer system based on Master-Slave architecture. It runs a parallel algorithm along with the Slave computers to process the collected EEG data in shorter period of time. Deterministic Finite Automata base recognizer named, DFAspike [14] has been incorporated in the parallel algorithm to recognize the ES present in EEG data.
-
II. Materials and Methods
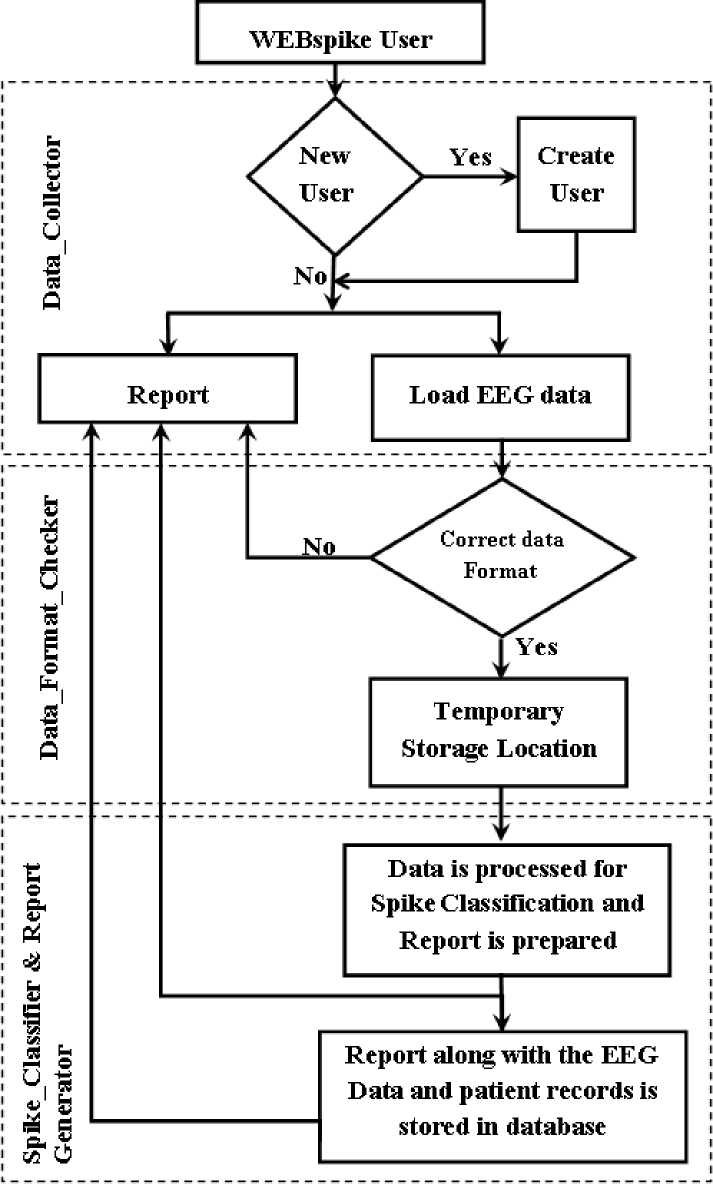
In the current work, a web-based system has been proposed that collects the EEG data from users residing over the globe and checks for presence of ES pattern in it. Once the inspection gets over, relevant report regarding the presence of ES patterns in the submitted EEG is sent back to the concerned users. The design of the system is logically divided into three parts: (i) Data_Collector, (ii) Data_Format_Checker and (iii) Report_Generator; as presented in fig 1, which is termed as WEBspike. The complete functionality of the WEBspike is represented in the flowchart shown in fig 2.
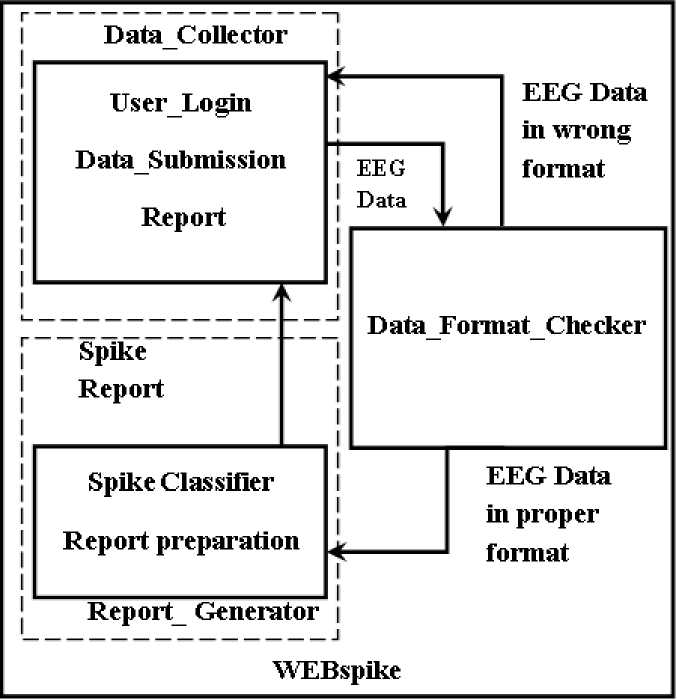
Fig. 1: Block diagram of WEBspike
New
User
Correct data Format ifi
Yes
Yes --->
Report along with the EEG Data and patient records is stored in database
Data is processed for Spike Classification and Report is prepared
Temporal^
Storage Location
Load EEG data
WEBspike User
Report
Create
User
Fig. 2: The flow chart that represent the functionality of WEBspike
Design of Data_Collector: It is a web site designed using J2EE. It contains two main pages. The first page is the Homepage, where a user can login to the system as shown in fig 3. In case of new user, there is a link ‘New User’ that provides facility to create a new account. Here the general details of the user are asked, among them the e-mail id is a compulsory field. A unique identification number (UIN) for the user is generated. The fetched details of the user, along with the UIN are stored as a record in a table named ‘WS_User_detail’ of a database named ‘Web_Spike_Database’. After being logged into the system, a new page is opened. The design of the page contains two basic modules ‘Submit_new_data’, and ‘Previously_submitted_data’ as shown in fig 4.
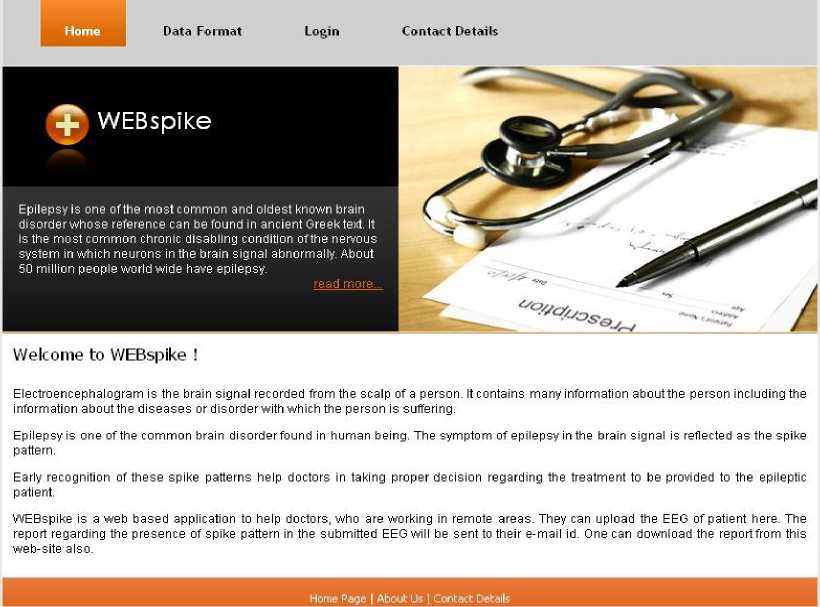
Fig. 3: The homepage of the proposed web application WEBspike
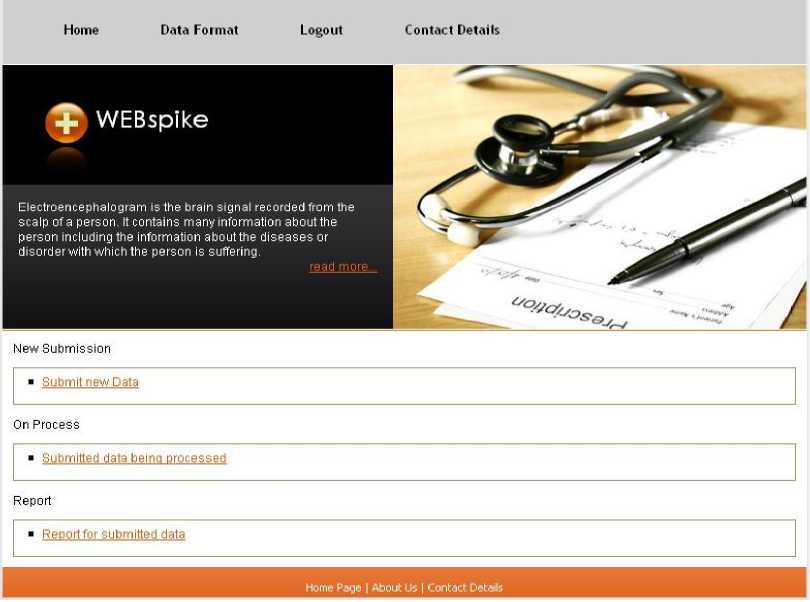
Fig. 4: The user account page appears on login to the proposed web application WEBspike
In the ‘Submit_new_data’ module, the user can upload the digitized EEG data stored in text file with file extension .DAT or .TXT. It also asks the detail of the patient and the recording details to upload. The uploaded data along with the patient detail is then transferred to the ‘Data_Format_Checker’ module for uploaded data file format verification.
The ‘Previously_submitted_Data’ module contains two sub modules: ‘On Process’ and ‘Report’. The already submitted data, which is being processed, are shown in the ‘On Process’ part and ‘Report’ part contains the detail of the data, which has been submitted by the same user sometime in the past with the corresponding report generated by the ‘Report_Generator’ module for the same data. The user can download the report at any time by login to the system. The same report is also e-mailed to the user.
-
2.2 Functionality of the Data_Format_Checker:
This module of WEBspike, checks for the proper format of the uploaded EEG data file. It is well known that each specialized bio-signal recorder creates the files in a specific file format, which is compatible only with that particular machine. Consideration of all the file formats for this web-based application is very difficult task. Therefore, for the purpose of global acceptability, the current proposed system has been intentionally designed for processing only the text files with extension .DAT or .TXT. The file must contains the EEG data in form of two-dimensional matrix that have two column. The data in the first column represent the time values when the EEG sample is recorded and the second column contains the EEG amplitude values recorded at that particular time point. The detail of the file type is also given in help page of the website.
If the uploaded EEG data files are found in the required format then they are stored in a table named ‘Patient_EEG_Data’ of the database
‘Web_Spike_Database’ as a record. A directory is created at the location ‘C:\WEBspike\PatientData\’ of the Master computer. This location temporarily stores the data and the reports of the patient for interprocessing. Once the processing of data of a patient is completed, the corresponding directory is automatically removed. If more than one user say ‘n’ number of users are logged simultaneously and uploading the patients EEG data file, then ‘n’ numbers of directories will be created. The directory name consists of two strings:
-
• The first three symbols of it is a three digit number ‘x’, which is equal to the instant id number of the user. Instant id number is assigned to each user who have logged for uploading the patient data.
-
• Next, ten symbols represents the UIN of each logged user.
As soon as, the EEG data is kept in a directory, the xth flag of the ‘Data_Ready_Flag’ Vector (DRFV) is set to represent that the xth directory is ready with the EEG data.
If the uploaded files do not match, the validity criteria, then with proper message, the files are sent back to the users via e-mail suggesting to upload the data again with proper format. The Data format tab in the home page contains the detail of the EEG file format that is supported by the proposed system.
This module of WEBspike is a multicomputer that is designed with Master-Slave architecture as shown in fig 5. It is having five computer. One among them is treated as Master and remaining four computers are treated as the Slave Computers. All the computers in a multicomputer architecture can communicate with each other. But in this particular application, the intercommunication among the Slave Computers are not needed and hence, in the block diagram of the Multicomputer, no connection among the Slave computers has been shown. The Master computer is having two tasks:
-
• It checks the status of DRFV in a regular interval. As soon as it finds that any flag of the DRFV is set, it takes the EEG data files from the corresponding directory and distributes it among the Slave Computers.
-
• It collects the details of the recognized spike patterns sent by the Slave Computers and prepares a report. Further, it stores the report in the same directory, from where it had taken the EEG data files. To indicate the ‘Data_Collector’ module about the ready report, it sets the xth flag of ‘Report_Ready_Flag’ Vector (RRFV) and reset the xth flag of DRFV.
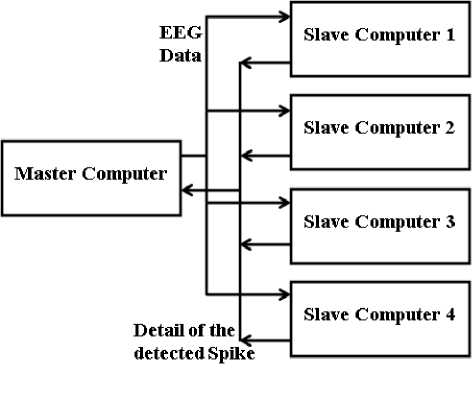
Fig. 5: Block diagram of the multicomputer used in WEBspike
To achieve the above-mentioned tasks, this module runs a parallel program. The parallel algorithm SPIKE_FINDER( ) and it functionality have already been proposed by the author in earlier work [15]. In the current work, the same algorithm has been used with the following three required modification:
-
a) In the earlier suggested parallel model of EEG data processing, the Master Computer was reading EEG file and distributing the fractions of EEG data among the Slave Computers in overlapped fashion. In the current work, the Master Computer will distribute the EEG data files among the Slave Computers, i.e. , a Slave Computer will get whole EEG data file at a time.
-
b) In the earlier design, the computers were divided into three logical group: Master, Intermediate and Result Accumulator with one Master and one Result Accumulator computer. The Master was dedicated to distribute the data among the Intermediate Computers and the Result Accumulator Computer was used to accumulate the results gathered from the Intermediate Computers. In this design, there is only one Master Computer and rest are Slaves. The role of the Master Computer is not only to distribute the EEG data files among the Slaves, but also to accumulate results obtained by the Slave Computers. The execution model of the algorithm is shown in fig 6. It is same as a typical parallel algorithm written in Message Passing Interface (MPI).
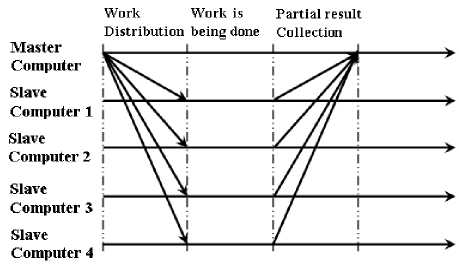
Fig. 6: The execution model of a basic MPI program
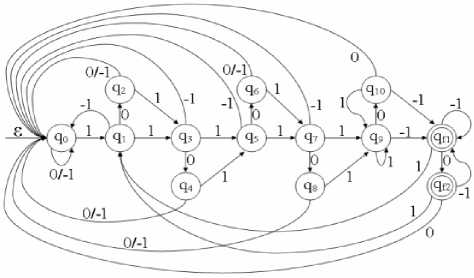
Fig. 7: Pictorial representation of the transition function used for modelling the optimized system DFAspike to recognize the Epileptic spike patterns
-
c) The earlier design was using Deterministic Finite Automata (DFA) based algorithm named DFASR for
-
2.4 Sending the report to the user:
-
2.5 Message Passing Interface (MPI):
ES recognition [13]. The DFASR was having an average accuracy of 95.47% with percentage misclassification 4.63%. In the current proposed web based model of EEG data processing, for ES recognition in EEG data, another DFA based algorithm named DFAspike [14] as shown in fig 7 has been used, which works with higher accuracy (99.09%) and very less misclassification percentage, i.e., 0.91%. In addition to it, the DFAspike algorithm is less complex than the DFASR.
The current version of parallel algorithm, used in WEBspike as spike recognizer has been named as ‘PARALLEL_SPIKE_FINDER_WITH_DFAspike’ and is given in ‘Appendix – I’. In the current proposed work, the multicomputer system contains only four Slave Computers, but the proposed algorithm can be used in a system with ‘n’ number of Slave Computers.
The ‘Data_Collector’ checks the RRFV regularly. As soon as it finds any flag of RRFV set, it take the generated report and mails it to the e-mail id of the corresponding user and load the report to its ‘Previously_submitted_data’ module. Further, it stores the report in ‘Patient_EEG_Data’ table as a field of the record of corresponding patient. Once the report is placed properly, and sent to the user, it deletes the directory created by the ‘Data_Format_Checker’ for that user in location ‘C:\WEBspike\PatientData\’.
The parallel algorithm is written in MPI. It is a standard for inter-process communication on distributed-memory multiprocessor. A broadly based group of parallel computer vendors, government laboratories, and universities [17] designed the standard. Implementation of the standard is usually left up to the designers of the systems on which MPI runs, but a public domain implementation, MPICH, is available [18]. It is a set of library routines for C/C++ and FORTRAN. The MPI is a standard interface, so code written for one system can easily be ported to another system with those libraries. It can be used to write programs for efficient execution on a wide variety of parallel machines, including massively parallel supercomputers, shared-memory multiprocessors and networks of workstations.
-
III. Results and Discussion
Webspike is a web-based application to help doctors and clinicians working in remote areas and not having smart tools for detection of ES in recorded EEG. For recognition of ES pattern in the input EEG data, it uses the earlier proposed algorithm DFAspike. For testing the application, ten users login id were created and 10
EEG data files were loaded through each login id. The comparative report generated by the WEBspike with respect to manual inspection is shown in fig 8. The recognition rate of ES present in EEG was achieved as 99.09% with the average misclassification of 0.09%.
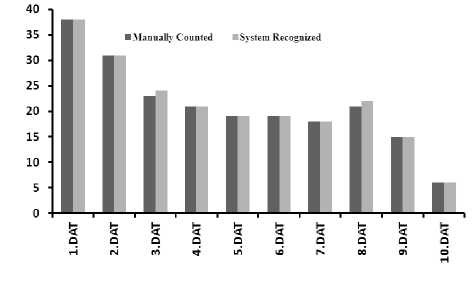
Fig. 8: Comparison of Spikes counted manually and by using WEBspike
Since this system will be made available in the internet, a number of users can upload the EEG data of patients to get instant analysis at a time. The algorithm used in this work currently supports only one thousand users to upload the EEG data files at a time, but this number can be increased by making few modifications in the program.
The total number of EEG files uploaded by different users simultaneously can be big enough and hence accumulating huge size EEG data to be analyzed. Any sequential algorithm may fail to handle such enormous data. Therefore, a Master-Slave architecture based multicomputer has been used in the server side that runs the parallel algorithm, making multiple numbers of computers to work together to analyze such huge data in shorter span of time. For the experimentation purpose, four Slave Computers have been used in the proposed system, but the algorithm used in it for analysis of EEG for ES is a generalized one that supports the scalability, i.e., by making small changes in the program, it can be made to work with more than four Slave Computers for the real application. Thus, the designed system is completely scalable.
For recognition of ES patterns in the EEG data, in the current work, an optimized DFA based algorithm DFAspike has been used. There are two advantages of using it:
-
• Its recognition rate with the tested EEG data with ES is very high, i.e. , 99.09%. This result can be
compared with other available system. It promises to produce result that is more accurate as far as the recognition of ES in EEG is concerned.
-
• It is suggested that the DFA based algorithms are one among the most suitable tool for applying in real time system, because they take only two contiguous EEG data points at time to process, hence buffering of a large size data is not needed at all. Thus, it needs very less memory to work with and hence, improve the overall efficiency of the system.
However, right now this system supports only the text files with extension name .DAT or .TXT for the EEG data to be stored and processed. These files should contain the EEG data in two dimensional array formats having only two columns, the first column contains the time series when the sample of EEG data is recorded and the second column contains the EEG sample in form of electric potential taken at a particular time. Thus, each row of the file contains only one time value and one EEG amplitude value separated by a blank space or a tab or any other separating character. The algorithm used in this work supports the processing of the EEG data recorded with a sampling rate of 256 samples per second. But, it can be made to work with a range of different sampling rates of EEG recording. Further, the current system processes single channel EEG recording data that can be modified to process multi-channel EEG data.
-
IV. Conclusion
The ‘World Wide Web’ has broken the geographical boundary of data processing and communication. A number of web-based applications are available, which are working successfully and providing variety of services to the internet users. In the current work, a web-based application has been proposed that facilitates the internet users especially doctors, clinicians and pathologists to analyze the EEG data of an epileptic patient for ES patterns beyond the geographical boundary. Further, such applications can be designed for analysis of other bio-signals as well as for medical images to ease the diagnostic complexities in the area of medical science.
Список литературы WEBspike: A New Proposition of Deterministic Finite Automata and Parallel Algorithm Based Web Application for EEG Spike Recognition
- Sinha RK. Electroencephalogram disturbances in different sleep-wake states following exposure to high environmental heat. Med. Biol. Eng. Computing (IEE) 2004;42:282 – 87.
- Adeli H, Zhou Z, Dadmehr N. Analysis of EEG records in an epileptic patient using wavelet transform. J. Neurosci. Methods. 2003;123: 69 – 87.
- Khan YU, Gotman J. Wavelet based automatic seizure detection in intracerebral electroencephalogram. Clin. Neurophysiol. 2003;114: 898 – 908.
- Zarjam P, Mesbah M, Boashash B. Detection of newborns EEG seizure using optimal features based on discrete wavelet transform. Proceeding of the IEEE International Conference on Acoustics Speech and Signal Processing. 2003;2: 265 – 268.
- Subasi A. Epileptic seizure detection using dynamic wavelet network. Expert Syst. Appl. 2005;29: 343 – 55.
- Subasi A. EEG signal classification using wavelet feature extraction and a mixture of expert model. Expert Syst. Appl. 2007;32: 1084 – 93.
- Indiradevi KP, Elias E, Sathidevi PS, Nayak SD Radhakrishnan KA. multi-level wavelet approach for automatic detection of epileptic spikes in the electroencephalogram. Computers in Biology and Medicine. 2008;38: 805 – 16.
- Ocak H. Automatic detection of epileptic seizures in EEG using discrete wavelet transform and approximate entropy. Expert Systems with Applications. 2009; 36:2027–36.
- Kiymik MK, Akin M, Subasi A. Automatic recognition of alertness level by using wavelets transform and artificial neural network. J. Neurosci. Methods. 2004;139: 231 – 40.
- Sinha RK, Ray AK, Agrawal NK. An artificial neural network to detect EEG seizures. Neurology India. 2004;52: 399-400.
- Subasi A. Automatic detection of epileptic seizure using dynamic fuzzy neural networks. Expert Syst. Appl. 2006;31: 320 – 28.
- Inan ZH, Kuntalp M. A study on fuzzy C-means clustering-based systems in automatic spike detection. Computers in Biology and Medicine 37: 2007;1160 – 66.
- Keshri AK, Sinha RK, Hatwal R, Das BN. Epileptic Spike Recognition in Electroencephalogram using Deterministic Finite Automata. Journal of Medical Systems 2009;33: 173 – 79.
- Keshri AK, Sinha RK, Singh A, Das BN. DFASpike: A new computational proposition for efficient recognition of Epileptic spike in EEG. Comput Biol Med. 2011;41 : 559 – 64.
- Keshri AK, Sinha RK, Mallick DK, Das BN., Parallel Algorithm to Analyze the Brain Signals : Application on Epileptic Spikes. J. Med. Syst. 2011;35:93 – 104.
- Fritschy J, Horesh L, Holder D, Bayford R. Applications of GRID in clinical neurophysiology and Electrical Impedance Tomography of brain function. Stud Health Technol Inform. 2005;112:138-45.
- Butler R, Lusk E. Monitors, messages, and clusters: the p4 parallel programming system. Journal of Parallel Computing. 1994;20(4): 547 – 64.
- Bala V, Kipnis S, Rudolph L, Snir M. Designing efficient, scalable, and portable communication libraries. SIAM 1993 Conference on Parallel Processing for Scientific Computing. 1993;862 – 72.
- Ktonas PY, Smith JR. Quantification of abnormal EEG characteristics. Comput Biol Med. 1974;4: 157 – 63.
- Adeli H, Dastidar SG, Dadmehr N. A Wavelet-Chaos Methodology for Analysis of EEGs and EEG Subbands to Detect Seizure and Epilepsy. IEEE Trans. on BME. 2007;54: 205- 11.


