Жирный хвост у овец: методы изучения генетических механизмов формирования фенотипа и идентифицированные гены-кандидаты (обзор)
Автор: Денискова Т.Е., Kunz E., Medugorac I., Доцев А.В., Brem G., Зиновьева Н.А.
Журнал: Сельскохозяйственная биология @agrobiology
Рубрика: Обзоры
Статья в выпуске: 6 т.54, 2019 года.
Бесплатный доступ
Жирнохвостые овцы, на долю которых приходится 25 % мировой популяции овец, широко распространены в странах Африки, Азии (A. Davidson, 1999), на Ближнем Востоке (S.P. Alves c соавт., 2013), а также в России (И.М. Дунин с соавт., 2013). Курдючный овечий жир был важным ингредиентом национальной кухни у многих этнических групп (С. Perry, 1995; A. Hajihosseinlo с соавт., 2015). В настоящее время растет спрос на постную пищу и снижение потребления жира, в связи с чем преимущества повышенного жироотложения в области хвостов у овец теряют значимость для производства продуктов питания (A. Nejati-Javaremi c соавт., 2007; M. Moradi c соавт., 2012). Развитие технологий геномного редактирования (N.A. Zinovieva с соавт., 2018) делает актуальным поиск генов, обусловливающих фенотип жирного хвоста, для их последующего нокаута при сохранении других ценных свойств жирнохвостых пород. В настоящем обзоре обобщены результаты исследований по идентификации генов-кандидатов, ассоциированных с признаком жирного хвоста. Для идентификации генов-кандидатов используются различные методы: поиск локусов под давлением селекции (signatures of selection) на основе расчета различий в частотах аллелей (значения Fst) или гаплотипов между популяциями (метод hapFLK) (M.H. Moradi c соавт., 2012; M.I. Fariello c соавт., 2013; C.M. Rochus c соавт., 2018); полногеномные ассоциативные исследования (genome-wide association study, GWAS), для проведения которых необходимо создать базу фенотипической вариабельности изучаемого признака (S.S. Xu c соавт., 2017); анализ вариации числа копий (copy number variation, CNV), способных изменять экспрессию генов из-за делеции или дублирования генов в регионах вариации (C. Zhu c соавт., 2016; Q. Ma c соавт., 2017; V. Bhanuprakash c соавт., 2018); изучение экспрессии генов с помощью технологии RNA-seq (RNA sequencing), основанной на транскриптомном анализе с использованием технологии секвенирования нового поколения (next generation sequencing, NGS) (W.A. Hoeijmakers, 2013). Суммируя результаты исследований, можно выделить наиболее значимые гены-кандидаты, ассоциированные с депонированием жира в хвостовой области у овец: BMP2 и VRTN (Z. Yuan c соавт., 2017; S. Mastrangelo c соавт., 2018; Z. Pan c соавт., 2019); PDGFD (C. Wei c соавт., 2015; S. Mastrangelo c соавт., 2018); гены семейства Homeobox (D. Kang c соавт., 2017; A.A. Yurchenko c соавт., 2019; A. Ahbara c соавт., 2019); SP9 (Z. Yuan c соавт., 2017; D. Kang c соавт., 2017); WDR92 и ETAA1 (Z. Yuan c соавт., 2017; L. Ma c соавт., 2018); CREB1 (S.S. Xu c соавт., 2017; L. Ma c соавт., 2018); FABP4 (M.R. Bakhtiarizadeh c соавт. 2013; B. Li c соавт., 2018); PPARA, RXRA, KLF11, ADD1, FASN, PPP1CA и PDGFA (C. Zhu c соавт., 2016; Q. Ma c соавт., 2017). Для поиска генов-кандидатов, вовлеченных в формирование жирного хвоста у российских пород, и последующего проведения полногеномных ассоциативных исследований заложена ресурсная популяция овец, полученная от скрещивания длинножирнохвостой карачаевской и короткотощехвостой романовской пород (ФНЦ животноводства - ВИЖ им. академика Л.К. Эрнста).
Домашние овцы, жирный хвост, курдюк, генетический маркер, днк-чипы
Короткий адрес: https://sciup.org/142226268
IDR: 142226268 | УДК: 636.38:636.082:577.21 | DOI: 10.15389/agrobiology.2019.6.1065rus
Текст обзорной статьи Жирный хвост у овец: методы изучения генетических механизмов формирования фенотипа и идентифицированные гены-кандидаты (обзор)
Выявление генов, ответственных за формирование хозяйственно полезных фенотипов и способных стать потенциальными мишенями для маркер-ориентированной и геномной селекции, а также геномного редактирования, — необходимый фундамент для развития генетических технологий в животноводстве (1). Изменившиеся требования человека к питанию, в частности возрастание спроса на постную пищу (2), обусловливают новые цели селекционно-племенной работы для ряда пород животных, включая овец. В мировой популяции овец 25 % составляют жирнохвостые и курдючные породы, которые широко распространены в странах Север- ной Африки, Азии (3, 4) и на Ближнем Востоке (5), а также в России (12 из 15 полугрубошерстных и грубошерстных пород овец России — жирнохвостые или курдючные) (6). Это связано с тем, что курдючный овечий жир был важным ингредиентом национальной кухни (7, 8). В настоящее время в мире, в том числе среди этнических групп юга России, наблюдается тенденция к снижению потребления жира, из-за чего созданные в течение тысячелетий породы овец с повышенным жироотложением теряют свою значимость для производства продуктов питания (2, 9).
Одомашнивание овец ( Ovis aries ) произошло 9000-11000 лет назад в регионе Плодородного полумесяца на территории современного Ирана (10). Фенотипический признак жирный хвост появился значительно позднее, чем тощий хвост. Во-первых, азиатский муфлон O. orientalis (вероятный предок домашних овец) (11-13) имеет короткий тощий хвост (дикий фенотип). То есть первые одомашненные овцы были короткотощехвостыми, а разведение овец с жирными хвостами началось на несколько тысячелетий позднее (14). Во-вторых, первые археологические упоминания о жирнохвостых овцах обнаружены в виде изображений на каменных чашах из Ура (Месопотамия), датируемых эпохой Урука III (3000-5000 лет назад) (14, 15).
Формирование жирного хвоста — аналога горба у верблюдов и зебувидного скота (9) с запасом ценного источника энергии стало следствием адаптации овец к неблагоприятным природным факторам: засухе, суровым зимам, нехватке корма и воды (16). Выносливые и неприхотливые овцы с жирными хвостами и курдюками были незаменимыми спутниками кочевых племен при длительных миграциях в Евразии и Африке (17, 18). Филогенетически (рис.) тип хвоста — это второй дифференцирующий фактор, что подтверждено результатами полногеномных исследований 25 российских пород овец (19).
Появление новых технологий, в частности геномного редактирования, позволяет направленно и точечно изменять геном овец (20-22). Нокаут генов, ассоциированных с признаком жирного хвоста, может стать эффективной стратегией, позволяющей элиминировать нежелательный фенотип при сохранении других ценных свойств жирнохвостых пород, таких как адаптационные качества, устойчивость к болезням, размеры туловища, качества мяса. В связи с этим важна идентификация причинных мутаций, определяющих депонирование жира в хвостовой области у овец.
В настоящем обзоре обобщены и проанализированы результаты исследований генетической обусловленности признака жирного хвоста у пород овец из разных стран мира, в том числе с указанием идентифицированных генов-кандидатов, вовлеченных в его формирование. Впервые проведен детальный анализ применяемых методических и биоинформаци-онных подходов для изучения генетических аспектов депонирования жира в хвостовой части у овец.
Поиск локусов, находящихся под давлением селекции (signatures of selection, selective sweeps). Доместикация и длительная селекция по хозяйственно полезным признакам (настриг и качество шерсти, мясная и молочная продуктивность, качество овчин), а также необходимость адаптации к новым условиям разведения (климат, кормовые ресурсы, системы содержания) значительно преобразовали геномы современных пород овец, оставляя после себя так называемые отпечатки селекции (23). Поиск регионов, находящихся под давлением селекции, — это один из наиболее популярных подходов для идентификации генов-кандидатов и картирования локусов количественных признаков (quantita- tive trait loci, QTL). Этот подход представляет собой не только инструмент для изучения эволюционной истории популяций и анализа адаптивных мутаций (23), но и имеет важное прикладное значение. Так, за последние 10 лет у овец идентифицированы гены-кандидаты, достоверно ассоциированные с пигментацией кожи (24-26), наращиванием мышечной массы (27), молочной продуктивностью (28), репродуктивными свойствами (25, 29), качеством шерсти (30), устойчивостью к паразитам (31), а также с формированием рогатых или безрогих фенотипов (25, 29).
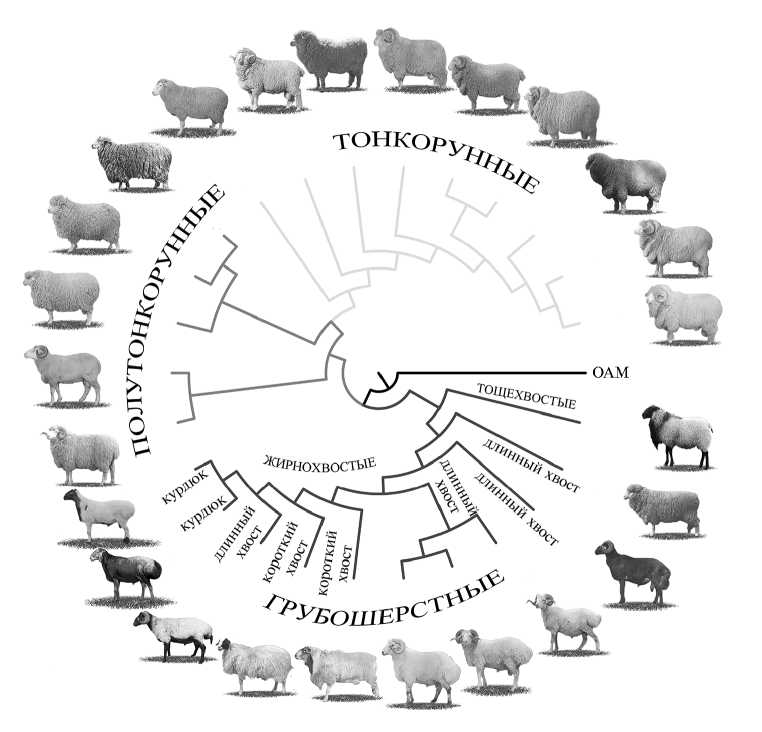
Филогенетическое дерево российских пород овец ( Ovis aries ) в зависимости от типа шерсти и типа хвоста. Используемые фотографии взяты из справочника пород и типов сельскохозяйственных животных, разводимых в Российской Федерации (6). OAM — Ovis ammon (argali), группа, которая была взята для укоренения дерева (outgroup).
Для поиска геномных регионов, находящихся под давлением селекции, применяется несколько методик биоинформационного анализа, основанных на установлении различий в частотах аллелей (расчет значений показателя F st ) (32, 33) или в частотах гаплотипов между популяциями (метод hapFLK) (34).
Первые попытки поиска генов-кандидатов, ассоциированных с жироотложением в области хвоста, начались с появлением ДНК-чипа OvineSNP50 BeadChip («Illumina, Inc.», США) со средней полностью покрытия генома (∼ 50 тыс. SNP, single nucleotide polymorphism). На его основе Международный консорциум по геномике овец (International Sheep Genomics Consortium, ISGC) в рамках проекта Ovine HapMap провел гено- типирование 2819 овец 74 пород и открыл доступ к cгенерированным SNP-профилям (25). В 2012 году M.H. Moradi с соавт. (9) провели сравнительный скрининг данных полногеномного генотипирования овец с тощими и жирными хвостами из числа местных иранских пород и овец, включенных в проект Ovine HapMap. В результате сканирования на X-, 5-й и 7-й хромосомах идентифицировали три геномных региона, находящихся под давлением отбора. Показано, что высокая степень гомозиготности в регионах на Х- и 5-й хромосомах способствует образованию жирного хвоста, а на 7-й хромосоме — обусловливает развитие тощего хвоста.
B. Moioli с соавт. (35) исследовали группы, включающие тощехвостых итальянских овец и две единственные европейские жирнохвостые породы (латичауда и киприотская жирнохвостая). Были обнаружены гены BMP2 и VRTN — по мнению авторов, наиболее вероятные кандидаты, ответственные за регуляцию процессов жироотложения в хвостовой области у овец. Так, костный морфогенетический белок 2 ( BMP2 ) играет важную роль в развитии костей и хрящей, а вертин ( VRTN ) отвечает за изменение числа позвонков (36). Ген BMP2 также находился под давлением селекции у местных египетских (37) и китайских жирнохвостых овец (38). Используя чип Ovine SNP50 BeadChip, C. Wei c соавт. (4) в качестве возможного кандидата предложили ген PDGFD , вовлеченный в дифференцировку преадипоцитов и характеризующийся высокой экспрессией в жировых тканях. Недавние исследования подтвердили роль генов BMP2 , VRTN и PDGFD как в формировании жирного хвоста, так и в развитии приспособленности к климату пустынь (39, 40).
В результате исследования местных китайских жирнохвостых пород овец Z. Yuan с соавт. (41) обнаружили 40 потенциальных генов-кандидатов, наиболее значимые из которых HOXA11 , BMP2 , PPP1CC , SP3 , SP9 , WDR92 , PROKR1 и ETAA1. Следует отметить, что гены PROKR1 и ETAA1 вовлечены в контролирование ожирения у людей (42, 43) и, вероятно, связаны с формированием жирных хвостов у овец.
Гены семейства Homeobox играют важную роль в процессах развития и морфологии скелета, крестца и хвоста (44). Предполагается, что гены HOXA11-13 , регулирующие число копчиковых позвонков, ответственны скорее за длину хвоста, чем за депонирование около него жира. Интересно, что ген HOXA11 был идентифицирован ранее, но не отнесен к группе кандидатов, ассоциированных с длиной или размерами хвоста (26) . С помощью ДНК-чипа высокой плотности Ovine Infinium® HD SNP BeadChip («Illumina, Inc.», США) ( ~ 600 тыс. SNP) показано, что гены семейства Homeobox находились под давлением селекции в российских жирнохвостых породах овец: группа генов HOXC на 3-й хромосоме — в лезгинской, эдильбаевской и каракульской, группа генов HOXA на 4-й хромосоме — в карачаевской и буубэй (45).
Изучая местных эфиопских и ливийских жирнохвостых овец с помощью чипа OvineSNP50 Beadchip, A. Ahbara с соавт. (46) выявили гены ALX4 , HOXB13 , BMP4 , ассоциированные с ростом и развитием конечностей, скелета и формированием хвоста. При дальнейшем полногеномном секвенировании нового поколения (next generation sequencing, NGS) с глубоким покрытием у представителей этих пород был обнаружен сильный сигнал селекции в районе гена HOXB13 , что подтвердило его влияние на формирование типов хвостов у овец (47).
Для идентификации генов-кандидатов, ответственных за короткий жирный хвост, D. Zhi с соавт. (48) провели секвенирование полного гено-1068
ма овец китайской местной породы Хулунбуир (Hulunbuir). Несмотря на противоречивые результаты, было показано, что мутация c.G334T в гене T , регулирующем процессы развития позвонков, влияет на формирование короткого жирного хвоста у овец, что частично подтверждается проведенными ранее исследованиям (49). Кроме того, в результате изучения приспособленности жирнохвостых пород овец к засушливым климатическим условиям Египта обнаружили 172 потенциальных гена-кандидата, так или иначе вовлеченных в физиологические механизмы адаптации и регулирование морфологии тела и отдельных его частей, в том числе жирного хвоста (37, 50).
П олно ге но мные ас с оциативны е ис след ов ания (genome-wide association study, GWAS). Полногеномные ассоциативные исследования — мощный биоинформационный инструмент для идентификации геномных вариаций, связанных с количественными признаками у сельскохозяйственных животных (51), в том числе у овец (52, 53). В отличие от исследований по поиску регионов, находящихся под давлением селекции, для проведения GWAS необходимо наличие базы фенотипической изменчивости по интересующему признаку.
На основе сбора информации о промерах и генотипирования с помощью ДНК-чипа высокой плотности S. Xu с соавт. (54) выявили несколько SNP, ассоциированных с развитием жирного хвоста, которые были локализованы в генах, ответственных за липидный обмен ( CREB1 , STEAP4 , CTBP1 и RIP140 ). Тем не менее не был идентифицирован ни один из группы генов, предложенных ранее в качестве функциональных кандидатов (35, 41). Выдвинуто предположение о том, что область генома между 8889 Мb на X-хромосоме содержит ряд значимых SNP и служит потенциальным регионом, связанным с формированием жирного хвоста у овец (54).
Анализ вар иации числа копий (copy number variation, CNV). Анализ вариации числа копий — это биоинформационный подход для детекции генов-кандидатов и идентификации QTL, а также для изучения эволюционных механизмов одомашнивания животных и их приспособленности к различным условиям окружающей среды (55). Феномен CNV заключается в том, что некоторые участки генома размером от одной тысячи до миллионов пар оснований присутствуют в нескольких копиях, число которых варьируется у особей внутри популяции (55). Вариация числа копий — важный источник генетической изменчивости у особи, поскольку CNV способны изменять экспрессию генов и, соответственно, фенотип из-за делеции или дупликации генов в регионах вариации (56-58).
Впервые вариация числа копий у овец была изучена в 2011 году. При этом обнаружили только 135 регионов CNV, что, возможно, связано с несовершенством выбранного метода гибридизации посредством кроссамплификации с геномом крупного рогатого скота (59). В 2016 году с помощью специально разработанной платформы Roche NimbleGen 2.1M CGH («Roche NimbleGen, Inc.», США) и с применением новых методов верификации CNV у овец выявили около 3488 аутосомных регионов CNV (60). Кроме того, осуществлялись попытки поиска функциональных генов, попадающих в регионы CNV на основании данных анализа с ДНК-чипом средней плотности (61, 62). C. Zhu с соавт. (63), применив ДНК-чип высокой плотности, выявили гены-кандидаты, полностью перекрывающиеся в регионах CNV и ассоциированные с депонированием жира в хвосте у овец местных китайских пород — Хан (Han) жирнохвостая, Алтай (Altay) и тибетская, включая гены PPARA , RXRA , KLF11, ADD1 , FASN , PPP1CA и
PDGFA . Позже было установлено, что перечисленные гены перекрывались в регионе CNV у еще одной китайской местной породы — Тан (Tan) (64).
Транскриптомный анализ на основе NGS (next generation sequencing). Развитие методов секвенирования нового поколения (NGS) привело к созданию технологии RNA-seq (RNA sequencing), что вывело исследования экспрессии генов на новый уровень (65). Популярность подхода RNA-seq обусловлена рядом преимуществ, включая высочайшую разрешающую способность, отсутствие необходимости предварительного секвенирования изучаемой нуклеотидной последовательности, а также возможность повторного анализа данных, полученных с помощью RNA-seq, в случае появления более актуальной сборки генома (65-67).
На основе метода RNA-seq были идентифицированы гены NELL1 и FMO3 , которые, вероятно, вовлечены в регуляцию метаболизма жиров в жировых тканях (68). Избыточная экспрессия гена NELL1 может быть ключевым фактором уменьшения жировых отложений в хвостовой части (68). Дальнейшие исследования местных китайских пород овец (69, 70) позволили идентифицировать гены-кандидаты, играющие значимую роль в повышенном отложении жира в области хвоста, — SP9 и гены семейства Homeobox : HOXC11 , HOXC12 , HOXC13 (69), CREB1 , WDR92 и ETAA1 (70). Кроме того, по мнению L. Ma с соавт. (71), гены FMO2 , PLIN2 , PLIN3 , LEPR , PENK , ELOVL3 , ELOVL5 , PDK4 и SLC22A4 играют ключевую роль в депонировании жира, адипогенезе и биосинтезе жирных кислот .
Транскриптомный анализ китайских пород продолжили B. Li с со-авт. (72), которые исследовали генетическую регуляцию процессов липидного обмена у овец с жирными хвостами. Установлено, что FABP4 , ADI-POQ , FABP5 и CD36 — это наиболее часто транскрибируемые гены, связанные с депонированием жира у овец. Хорошо известна роль генов FABP4 и FABP5 в накоплении жира у крупного рогатого скота (73) и в формировании нежности мяса у овец (74). Гены CD36 и ADIPOQ служат транспортерами и регуляторами жирных кислот (75-77). Авторы отмечают, что гены FHC , FHC-псевдоген и ZC3H10 также могут быть вовлечены в регуляцию липидного обмена у овец (72). Предполагается связь гена ZNF395 с процессами адипогенеза (78).
Роль гена FABP4 в депонировании жира в области хвостов у некоторых пород овец обсуждалась и ранее, однако были получены весьма противоречивые результаты. Например, M.R. Bakhtiarizadeh с соавт. (79), определив степень экспрессии генов-кандидатов, в том числе FABP4 , FASN , SCD и LPL , обнаружили достоверно более высокую экспрессию гена FABP4 в жирных хвостах по сравнению с экспрессией в висцеральных жировых тканях (p < 0,05). Однако X. Ruixia с соавт. (80), отмечая высокую экспрессию гена FABP4 у курдючной породы Алтай, не нашли статистически достоверной разницы с контрольной группой, состоящей из овец с тощими хвостами. Окончательный вывод о роли гена FABP4 в формировании жирного хвоста и курдюка делать пока рано, поскольку перечисленные особенности могут быть породоспецифичными.
Существует предположение о регуляции экспрессии генов, связанных с отложением жира в хвостовой области у овец, с участием некодирующих РНК (long non-coding RNAs, lncRNAs) (81). Показана высокая корреляция экспрессии мРНК Lpin2 и Lpin3 с размерами жирных хвостов (82). Кроме того, установлена разница в экспрессии генов CPT1 (83) и OXCT1 (84) у жирнохвостых овец по сравнению с тощехвостыми, поэтому рассматриваемые гены можно считать вероятными кандидатами.
Информация об идентифицированных потенциальных генах-кандидатах обобщена в таблице.
Гены-кандидаты, вовлеченные в формирование фенотипа жирный хвост у овец (Ovis aries), и методы их идентификации
|
Ген 1 |
Хромосом |
а Метод |
^ Функции |
^ Ссылка |
|
ADIPOQ (адипонектин, adiponectin) |
1-я |
RNA-seq |
Окисления ЖК и глюкозы |
(72) |
|
LEPR (рецептор лептина, leptin receptor) |
1-я |
RNA-seq |
Депонирование жира, адипогенез |
(71) |
|
RIP140 (NRIP1) (взаимодействующий протеин 1 ядер-ного рецептора, nuclear receptor interacting protein 1) |
1-я |
GWAS |
Регуляции метаболизма липидов и глюкозы |
(54) |
|
CREB1 (cAMP-чувствительный элемент, связывающий протеин 1, cAMP responsive element binding protein 1) |
2-я |
RNA-seq GWAS |
Липидный обмен, гомеостаз глюкозы, дифференцировка адипоцитов |
(54, 70) |
|
LPL (липопротеин-липаза, lipoprotein lipase) |
2-я |
RNA-seq |
Высвобождение ЖК и глицерина посредством гидролиза триглицеридов |
(79) |
|
PLIN2 (перилипин 2, perilipin 2) |
2-я |
RNA-seq |
Депонирование жира |
(71) |
|
SP3 (Sp3-транскрипцион-ный фактор, Sp3 transcription factor) |
2-я |
SPS |
Ингибирование дифференцировки адипоцитов |
(41) |
|
SP9 (Sp9-транскрипцион-ный фактор, Sp9 transcription factor) |
2-я |
SPS, RNA-seq |
Жироотложение посредством прилипания мезенхимальных клеток к адипоцитам |
(41, 69) |
|
ZNF395 (белок цинковых пальцев, zinc finger protein 395) |
2-я |
RNA-seq |
Дифференцировка преадипоцитов, определение линии клеток-предшественников мезенхимы |
(78) |
|
SLC22A4 (член семейства SLC22A, SLC22A family member) |
3-я |
RNA-seq |
Энергетический обмен, депонирование жира |
(71) |
|
CPT1 (карнитин пальмито-ил-трансфераза, 1 carnitine palmitoyltransferase 1) |
3-я |
RNA-seq |
Участие в метаболизме ЖК в печени |
(83) |
|
ETAA1 (ассоциированный антиген 1 опухоли Юинга, Ewing tumor-associated antigen 1) |
3-я |
SPS, RNA-seq |
Распределение и депонирование жира в организме |
(41, 70) |
|
HOXC11 (гомеобокс С11-13, homeobox C11-13) |
3-я |
RNA-seq, SPS |
Нокаут приводит к трансформации позвонков |
(45, 69) |
|
KLF11 (Kruppel-подобный фактор 11, Kruppel-like factor 11) |
3-я |
CNV |
Фактор транскрипции бурого жира |
(64) |
|
PPARA (пероксиом проли-фератор-активируемый рецептор- α , peroxisome proli-ferator-activated receptor- α ) |
3-я |
CNV |
Коактиватор метаболизма ЖК |
(64) |
|
PROKR1 (рецептор прокине-тина 1, prokineticin receptor 1) |
3-я |
SPS |
Подавление пролиферации и дифференцировки преадипоцитов |
(41) |
|
RXRA (ретиноевый X рецептор A, retinoic X receptor A) |
3-я |
CNV |
Гомеостаз липидов |
(64) |
|
WDR92 (WD повтор домена 92, WD repeat domain 92) |
3-я |
SPS, RNA-seq |
Взаимодействие с фосфолипидами |
(41, 70) |
|
ZC3H10 (белок цинковых пальцев CCCH 10, zinc finger CCCH-type containing 10) |
3-я |
RNA-seq |
Гомеостаз адипоцитов. Расположен в QTL, связанном с количеством внутреннего жира. |
(72) |
|
HOXA11 (гомеобокс А11, homeobox A11) |
4-я |
SPS |
Регуляция вариации числа копчиковых позвонков |
(26, 41, 45) |
|
PDK4 (киназа пируватдегидрогеназы 4, pyruvate dehydrogenase kinase 4) |
4-я |
RNA-seq |
Депонирование жира, адипогенез |
(71) |
|
STEAP4 (металлоредуктаза STEAP4, STEAP4 metallore-ductase) |
4-я |
GWAS |
Кодирует металлоредуктазу в жировой ткани, участвующую в метаболизме адипоцитов |
(54) |
|
PLIN3 (перилипин 3, perilipin 3) |
5-я |
RNA-seq |
Депонирование жира |
(71) |
|
ADD1 (фактор детерминации и дифференциации адипоцитов 1, adipocyte determination and differentiation factor 1) |
6-я |
CNV |
Дифференцировка адипоцитов и гомеостаз холестерина |
(64) |
Продолжение таблицы
|
CTBP1 (белок 1, связываю- |
6-я |
GWAS |
Окисление ЖК; ингибирование приво- |
(54) |
|
щий С-конец, C-terminal binding protein 1) |
дит к ожирению печени |
|||
|
BMP4 (костный морфогенетический белок 4, bone morphogenetic protein 4) |
7-я |
SPS |
Рост и развитие конечностей, скелета и формирование хвоста |
(46) |
|
VRTN (вертин, ассоциированный с развитием позвонков, vertnin, vertebrae development associated) |
7-я |
SPS |
Изменение числа позвонков |
(35, 39, 40) |
|
T(TBXT) (эмбриональный транскрипционный фактор, гомолог brachybury, T-box transcription factor T, T/Brachybury) |
8-я |
SPS |
Развитие позвонков, формирование короткого жирного хвоста у овец |
(48, 49) |
|
FABP4 (связывающий жирные кислоты белок 4, fatty acid binding protein 4) |
9-я |
RNA-seq |
Доставка ЖК к местам аккумуляции или(72, 79, производства энергии. Высокая экс- 80) прессия увеличивает время дифференцировки адипоцитов |
|
|
FABP5 (связывающий жирные кислоты белок 5, fatty acid binding protein 5) |
9-я |
RNA-seq |
Компенсация потери FABP4 в адипоцитах |
(72) |
|
PENK (проэнкефалиновый А, proenkephalin) |
9-я |
RNA-seq |
Аккумуляция жира |
(71) |
|
FASN (синтаза жирных кислот, fatty acid synthase) |
11-я |
CNV |
Синтез ЖК de novo, отложение жира и анаболизм ЖК |
(64) |
|
HOXB13 (гомеобокс В13, homeobox B13) |
11-я |
SPS |
Рост и развитие конечностей, скелета и формирование хвоста |
(46, 47) |
|
FMO2 (флавинсодержащая диметиланилиновая монок-сигеназа 2, flavin containing dimethylaniline monoxygenase 2) |
12-я |
RNA-seq |
Депонирование жира, адипогенез и биосинтез ЖК |
(71) |
|
BMP2 (костный морфогенетический белок 2, bone morphogenetic protein 2) |
13-я |
SPS |
Развитие костей и хрящей |
(35, 3741) |
|
Lpin3 (липин 3, lipin 3) |
13-я |
RNA-seq |
Метаболизм липидов |
(82) |
|
ALX4 (ALX гомеобокс 4, ALX homeobox 4) |
15-я |
SPS |
Рост и развитие конечностей, скелета и формирование хвоста |
(46) |
|
PDGFD (тромбоцитарный фактор роста D, platelet-derived growth factor D) |
15-я |
SPS |
Ингибирование дифференцировки преадипоцитов |
(4, 39, 40) |
|
OXCT01 (3-оксокислота КоА-трансфераза 1, 3-oxoacid CoA-transferase 1) |
16-я |
RNA-seq |
Метаболизм кетоновых тел; нокдаун приводит к накоплению липидов в адипоцитах |
(84) |
|
PPP1CC (каталитическая субъединица гамма проте-инфосфатазы 1, protein phosphatase 1 catalytic subunit gamma) |
17-я |
SPS |
Дефосфорилирование и инактивация гликогенсинтазы в скелетных мышцах |
(41) |
|
ELOVL5 (элонгаза жирных кислот 5, fatty acid elongase 5) |
20-я |
RNA-seq |
Удлинение полиненасыщенных длинноцепочечных ЖК |
(71) |
|
FHC (тяжелая цепь ферритина, ferritin heavy chain) |
21-я |
RNA-seq |
Регуляция активности жировых клеток |
(72) |
|
FMO3 (флавиносодержащая монооксигеназа 3, flavin containing monooxygenase 3) |
21-я |
RNA-seq |
Ингибирует окисление ЖК |
(68) |
|
NELL1 (NEL-подобный нейронный EGFL 1: белок, сильно экспрессируемый в нервной ткани, кодирующий эпидермальный фактор роста, подобный домену 1, NEL-like 1 neural EGFL like 1, a protein strongly expressed in neural tissue encoding epidermal growth factor like domain) |
21-я |
RNA-seq |
Усиливает остеогенную дифференциацию и ослабляет дифференцировку жировой ткани |
(68) |
|
PPP1CA (каталитическая фосфатопротеина 1-каталитическая субъединица |
21-я |
CNV |
Превращает фосфорилазу А в фосфорилазу В |
(64) |
A, phosphoprotein phosphatase 1 catalytic subunit)
22-я RNA-seq
23-я RNA-seq
24-я CNV
ELOVL3 (элонгаза жирных кислот 3, ELOVL fatty acid elongase 3)
Lpin2 (липин 2, lipin 2)
PDGFA (тромбоцитарный фактор роста альфа, platelet-derived growth factor alpha)
П р и м е ч а н и е. RNA-seq — транскриптомный
Продолжение таблицы
Энергетический обмен, удлинение(71)
насыщенных и мононенасыщенных ЖК
Участвуют в метаболизме липидов(82)
Дифференцировка преадипоцитов(64)
анализ экспрессии генов на основе секвенирования
нового поколения (NGS), CNV — анализ вариации числа копий (copy number variation), SPS — поиск регионов и локусов, находящихся под давлением селекции (signatures of selection, selective sweeps), GWAS — полногеномные ассоциативные исследования (genome-wide associated study); ЖК — жирные кислоты.
Для поиска генов-кандидатов, ассоциированных с формированием жирного хвоста и курдюка у российских пород овец, заложена ресурсная популяция овец, полученная от скрещивания длинножирнохвостой карачаевской и короткотощехвостой романовской пород овец (ФНЦ животноводства — ВИЖ им. академика Л.К. Эрнста). У гибридов F 2 и возвратных кроссов будут сделаны промеры хвостов в динамике на 6-е, 42-е, 90-е и 180-е сут после рождения. Все животные ресурсной популяции будут гено-типированы с помощью ДНК-чипов высокой плотности с последующим проведением GWAS. Использование ресурсных популяций для картирования QTL имеет ряд преимуществ по сравнению с выбором случайных популяций, в частности снижается доля ложноположительных результатов (false discovery rate, FDR) и повышается точность картирования (85). В связи с этим выявленные гены-кандидаты, ассоциированные с жироотложением в хвостах и характерные для российских пород, станут возможными мишенями для геномного редактирования жирнохвостых овец с целью увеличения производства постной баранины.
Таким образом, для выявления потенциальных генов-кандидатов, ассоциированных с повышенным жироотложением в хвостовой области у овец, наиболее часто применяются поиск локусов под давлением селекции, полногеномные ассоциативные исследования, анализ вариации числа копий, изучение экспрессии генов на основе транскриптомного анализа. В применении этих подходов существуют достоинства и ограничения. Так, полногеномные ассоциативные исследования — высокоточный и эффективный метод картирования QTL, но для них требуется база по фенотипической изменчивости изучаемого хозяйственно полезного признака. Для поиска локусов под давлением селекции и анализа вариации числа копий информация о фенотипах исследуемых животных необязательна, но результаты, полученные с помощью обоих подходов, требуют валидации посредством количественной ПЦР амплификации или секвенирования идентифицированных участков генома. Транскриптомный анализ — высокоразрешающий методом, однако образцы органов и тканей для его проведения должны быть отобраны с учетом соблюдения жестких временных и температурных правил. К наиболее значимым генам-кандидатам, ассоциированным с депонированием жира в хвостовой области у овец и выявленным более чем в одной работе, относятся BMP2 и VRTN, PDGFD, гены семейства гомеобоксов, SP9, CREB1, PPARA, RXRA, KLF11, ADD1, FASN, PPP1CA и PDGFA. Несмотря на значительное количество исследований по выявлению генетических механизмов обусловленности фенотипа жирный хвост у овец, в настоящее время достоверно известно только то, что в его формирование вовлечено более одного гена. Для поиска генов-кандидатов у российских пород овец планируется провести полногеномное ассоциативное исследование ресурсной популяции овец, полу- ченной от скрещивания длинножирнохвостой карачаевской и короткотощехвостой романовской пород.
Список литературы Жирный хвост у овец: методы изучения генетических механизмов формирования фенотипа и идентифицированные гены-кандидаты (обзор)
- Белоус А.А., Сермягин А.А., Костюнина О.В., Брем Г., Зиновьева Н.А. Изучение генетической архитектуры конверсии корма у хряков (Sus scrofa) породы дюрок на основе полногеномного анализа SNP. Сельскохозяйственная биология, 2019, 54(4): 705-712 ( ). DOI: 10.15389/agrobiology.2019.4.705rus
- Nejati-Javaremi A., Izadi F., Rahmati G., Moradi M. Selection in fat-tailed sheep based on two traits of fat-tail and body weight versus single-trait total body weight. Int. J. Agri. Biol., 2007, 9(4): 645-648.
- Davidson A. The Oxford companion to food. Oxford University Press, Oxford, UK, 1999.
- Wei C., Wang H., Liu G., Wu M., Cao J., Liu Z., Liu R., Zhao F., Zhang L., Lu J., Liu C., Du L. Genome-wide analysis reveals population structure and selection in Chinese indigenous sheep breeds. BMC Genomics, 2015, 16: 194 ( ). DOI: 10.1186/s12864-015-1384-9
- Alves S.P., Bessa R.J.B., Quaresma M.A.G., Kilminster T., Scanlon T., Oldham C., Milton J., Greeff J., Almeida A.M. Does the fat tailed Damara ovine breed have a distinct lipid metabolism leading to a high concentration of branched chain fatty acids in tissues? PLoS ONE, 2013, 8(10): e77313 ( ). DOI: 10.1371/journal.pone.0077313
- Дунин И.М., Данкверт А.Г. Справочник пород и типов сельскохозяйственных животных, разводимых в Российской Федерации. М., 2013.
- Perry C. The fate of the tail. In: Disappearing foods: studies in food and dishes at risk: Proceedings of the Oxford symposium on food and cookery /H. Walker (ed.). Pro-spect Books, UK, 1995.
- Hajihosseinlo A., Jafari S., Ajdary M. The relationship of GH and LEP gene poly-morphisms with fat-tail measurements (fat-tail dimensions) in fat-tailed Makooei breed of Iranian sheep. Adv. Biomed. Res., 2015, 4: 172 ( ).
- DOI: 10.4103/2277-9175.163995
- Moradi M.H., Nejati-Javaremi A., Moradi-Shahrbabak M., Dodds K.G., McEwan J.C. Genomic scan of selective sweeps in thin and fat tail sheep breeds for identifying of candidate regions associated with fat deposition. BMC Genetics, 2012, 13: 10 ( ).
- DOI: 10.1186/1471-2156-13-10
- Zeder M.A. Domestication and early agriculture in the Mediterranean Basin: origins, diffusion, and impact. Proceedings of the National Academy of Sciences, 2008, 105(33): 11597-11604 ( ).
- DOI: 10.1073/pnas.0801317105
- Hiendleder S., Kaupe B., Wassmuth R., Janke A. Molecular analysis of wild and domestic sheep questions current nomenclature and provides evidence for domestication from two different subspecies. Proc. R. Soc. Lond. B., 2002, 269(1494): 893-904 ( ).
- DOI: 10.1098/rspb.2002.1975
- Rezaei H.R., Naderi S., Chintauan-Marquier I.C., Taberlet P., Virk A.T., Naghash H.R., Rioux D., Kaboli M., Luikart G., Pompanon F. Evolution and taxonomy of the wild species of the genus Ovis (Mammalia, Artiodactyla, Bovidae). Molecular Phylogenetics and Evolution, 2010, 54(2): 315-326 ( ).
- DOI: 10.1016/j.ympev.2009.10.037
- Demirci S., Koban Baştanlar E., Dağtaş N.D., Pişkin E., Engin A., Özer F., Yüncü E., Doğan S.A., Togan İ. Mitochondrial DNA diversity of modern, ancient and wild sheep (Ovis gmelinii anatolica) from Turkey: new insights on the evolutionary history of sheep. PLoS ONE, 2013, 8(12): e81952 ( ).
- DOI: 10.1371/journal.pone.0081952
- Ryder M.L. Sheep and man. Gerald Duckworth & Co. Ltd., London, 1983.
- Epstein H. The Awassi sheep with special reference to the improved dairy type. FAO Animal production and health paper, 1985, 57: 282.
- Kashan N.E.J., Azar G.H.M., Afzalzadeh A., Salehi A. Growth performance and carcass quality of fattening lambs from fat-tailed and tailed sheep breeds. Small Ruminant Research, 2005, 60(3): 267-271 ( ).
- DOI: 10.1016/j.smallrumres.2005.01.001
- Lv F.H., Peng W.F., Yang J., Zhao Y.X., Li W.R., Liu M.J., Ma Y.H., Zhao Q.J., Yang G.L., Wang F., Li J.Q., Liu Y.G., Shen Z.Q., Zhao S.G., Hehua E., Gorkhali N.A., Farhad Vahidi S.M., Muladno M., Naqvi A.N., Tabell J., Iso-Touru T., Bruford M.W., Kantanen J., Han J.L., Li M.H. Mitogenomic meta-analysis identifies two phases of migration in the history of Eastern Eurasian sheep. Molecular Biology and Evolution, 2015, 32(10): 2515-2533 ( ).
- DOI: 10.1093/molbev/msv139
- Zhao Y.X., Yang J., Lv F.H., Hu X.J., Xie X.L., Zhang M., Li W.R., Liu M.J., Wang Y.T., Li J.Q., Liu Y.G., Ren Y.L., Wang F., Hehua E., Kantanen J., Arjen Lenstra J., Han J.L., Li M.H. Genomic reconstruction of the history of native sheep reveals the peopling patterns of nomads and the expansion of early pastoralism in East Asia. Molecular Biology and Evolution, 2017, 34(9): 2380-2395 ( ).
- DOI: 10.1093/molbev/msx181
- Deniskova T.E., Dotsev A.V., Selionova M.I., Kunz E., Medugorac I., Reyer H., Wimmers K., Barbato M., Traspov A.A., Brem G., Zinovieva N.A. Population structure and genetic diversity of 25 Russian sheep breeds based on whole-genome genotyping. Genet. Sel. Evol., 2018, 50(1): 29 ( ).
- DOI: 10.1186/s12711-018-0399-5
- Proudfoot C., Carlson D.F., Huddart R., Long C.R., Pryor J.H., King T.J., Lillico S.G., Mileham A.J., McLaren D.G., Whitelaw C.B., Fahrenkrug S.C. Genome edited sheep and cattle. Transgenic Res., 2015, 24(1): 147-153 ( ).
- DOI: 10.1007/s11248-014-9832-x
- Zhou S., Yu H., Zhao X., Cai B., Ding Q., Huang Y., Li Y., Li Y., Niu Y., Lei A., Kou Q., Huang X., Petersen B., Ma B., Chen Y., Wang X. Generation of gene-edited sheep with a defined Booroola fecundity gene (FecBB) mutation in bone morphogenetic protein receptor type 1B (BMPR1B) via clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated (Cas) 9. Reproduction, Fertility and Development, 2018, 30(12): 1616-1621 ( ).
- DOI: 10.1071/RD18086
- Zinovieva N.A., Volkova N.A., Bagirov V.A. Genome editing: state of the art and application to animal husbandry. Biotekhnologiya, 2018, 34(3): 9-22 ( ).
- DOI: 10.21519/0234-2758-2018-34-3-9-22
- Rochus C.M., Tortereau F., Plisson-Petit F., Restoux G., Moreno-Romieux C., Tosser-Klopp G., Servin B. Revealing the selection history of adaptive loci using genome-wide scans for selection: an example from domestic sheep. BMC Genomics, 2018, 19(1): 71 ( ).
- DOI: 10.1186/s12864-018-4447-x
- García-Gámez E., Reverter A., Whan V., McWilliam S.M., Arranz J.J., Consortium ISG, Kijas J. Using regulatory and epistatic networks to extend the findings of a genome scan: identifying the gene drivers of pigmentation in merino sheep. PLoS ONE, 2011, 6(6): e21158 ( ).
- DOI: 10.1371/journal.pone.0021158
- Kijas J.W., Lenstra J.A., Hayes B., Boitard S., Porto Neto L.R., San Cristobal M., Servin B., McCulloch R., Whan V., Gietzen K., Paiva S., Barendse W., Ciani E., Raadsma H., McEwan J., Dalrymple B., International Sheep Genomics Consortium Members. Genome-wide analysis of the world's sheep breeds reveals high levels of historic mixture and strong recent selection. PLoS Biology, 2012, 10(2): e1001258 ( ).
- DOI: 10.1371/journal.pbio.1001258
- Fariello M.I., Servin B., Tosser-Klopp G., Rupp R., Moreno C., Consortium ISG, San Cristobal M., Boitard S. Selection signatures in worldwide sheep populations. PLoS ONE, 2014, 9(8): e103813 ( ).
- DOI: 10.1371/journal.pone.0103813
- Fleming-Waddell J.N., Olbricht G.R., Taxis T.M., White J.D., Vuocolo T., Craig B.A., Tellam R.L., Neary M.K., Cockett N.E., Bidwell C.A. Effect of DLK1 and RTL1 but not MEG3 or MEG8 on muscle gene expression in Callipyge lambs. PLoS ONE, 2009, 4(10): e7399 ( ).
- DOI: 10.1371/journal.pone.0007399
- Moioli B., Scatà M.C., Steri R., Napolitano F., Catillo G. Signatures of selection identify loci associated with milk yield in sheep. BMC Genet., 2013, 14: 76 ( ).
- DOI: 10.1186/1471-2156-14-76
- Liu Z.H., Ji Z.B., Wang G.Z., Chao T.L., Hou L., Wang J.M. Genome-wide analysis reveals signatures of selection for important traits in domestic sheep from different ecoregions. BMC Genomics, 2016, 17(1): 863 ( ).
- DOI: 10.1186/s12864-016-3212-2
- Demars J., Cano M., Drouilhet L., Plisson-Petit F., Bardou P., Fabre S., Servin B., Sarry J., Woloszyn F., Mulsant P., Foulquier D., Carrière F., Aletru M., Rodde N., Cauet S., Bouchez O., Pirson M., Tosser-Klopp G., Allain D. Genome-wide identification of the mutation underlying fleece variation and discriminating ancestral hairy species from modern woolly sheep. Molecular Biology and Evolution, 2017, 34(7): 1722-1729 ( ).
- DOI: 10.1093/molbev/msx114
- Mcrae K.M., McEwan J.C., Dodds K.G., Gemmell N.J. Signatures of selection in sheep bred for resistance or susceptibility to gastrointestinal nematodes. BMC Genomics, 2014, 15: 637 ( ).
- DOI: 10.1186/1471-2164-15-637
- Weir B.S., Cockerham C.C. Estimating F-statistics for the analysis of population-structure. Evolution, 1984, 38(6): 1358-1370 ( ).
- DOI: 10.2307/2408641
- Porto-Neto L.R., Lee S.H., Lee H.K., Gondro C. Detection of signatures of selection using FST. In: Methods in molecular biology (methods and protocols) /C. Gondro, J. van der Werf, B. Hayes (eds.). Humana Press, Totowa, NJ, 2013, vol. 1019: 423-436 ( ).
- DOI: 10.1007/978-1-62703-447-0_19
- Fariello M.I., Boitard S., Naya H., SanCristobal M., Servin B. Detecting signatures of selection through haplotype differentiation among hierarchically structured populations. Genetics, 2013, 193(3): 929-941 ( ).
- DOI: 10.1534/genetics.112.147231
- Moioli B., Pilla F., Ciani E. Signatures of selection identify loci associated with fat tail in sheep. Journal of Animal Science, 2015, 93(10): 4660-4669 ( ).
- DOI: 10.2527/jas.2015-9389
- Mikawa S., Sato S., Nii M., Morozumi T., Yoshioka G., Imaeda N., Yamaguchi T., Hayashi T., Awata T. Identification of a second gene associated with variation in vertebral number in domestic pigs. BMC Genet., 2011, 12: 5 ( ).
- DOI: 10.1186/1471-2156-12-5
- Kim E.S., Elbeltagy A.R., Aboul-Naga A.M., Rischkowsky B., Sayre B., Mwacharo J.M., Rothschild M.F. Multiple genomic signatures of selection in goats and sheep indigenous to a hot arid environment. Heredity, 2016, 116: 255-264 ( ).
- DOI: 10.1038/hdy.2015.94
- Pan Z., Li S., Liu Q., Wang Z., Zhou Z., Di R., An X., Miao B., Wang X., Hu W., Guo X., Lv S., Li F., Ding G., Chu M., Li Y. Rapid evolution of a retro-transposable hotspot of ovine genome underlies the alteration of BMP2 expression and development of fat tails. BMC Genomics, 2019, 20(1): 261 ( ).
- DOI: 10.1186/s12864-019-5620-6
- Mastrangelo S., Moioli B., Ahbara A., Latairish S., Portolano B., Pilla F, Ciani E. Genome-wide scan of fat-tail sheep identifies signals of selection for fat deposition and adaptation. Animal Production Science, 2018, 59(5): 835-848 ( ).
- DOI: 10.1071/AN17753
- Mastrangelo S., Bahbahani H., Moioli B., Ahbara A., Al Abri M., Almathen F., da Silva A., Belabdi I., Portolano B., Mwacharo J.M., Hanotte O., Pilla F., Ciani E. Novel and known signals of selection for fat deposition in domestic sheep breeds from Africa and Eurasia. PLoS ONE, 2019, 14(6): e0209632 ( ).
- DOI: 10.1371/journal.pone.0209632
- Yuan Z., Liu E., Liu Z., Kijas J.W., Zhu C., Hu S., Ma X., Zhang L., Du L., Wang H., Wei C. Selection signature analysis reveals genes associated with tail type in Chinese indigenous sheep. Anim. Genet., 2017, 48(1): 55-66 ( ).
- DOI: 10.1111/age.12477
- Szatkowski C., Vallet J., Dormishian M., Messaddeq N., Valet P., Boulberdaa M., Metzger D., Chambon P., Nebigil C.G. Prokineticin receptor 1 as a novel suppressor of preadipocyte proliferation and differentiation to control obesity. PLoS ONE, 2013, 8(12): e81175 ( ).
- DOI: 10.1371/journal.pone.0081175
- Liu C.T., Monda K.L., Taylor K.C. et al. Genome-wide association of body fat distribution in African ancestry populations suggests new loci. PLoS Genetics, 2013, 9(8): e1003681 ( ).
- DOI: 10.1371/journal.pgen.1003681
- Mallo M., Wellik D.M., Deschamps J. Hox genes and regional patterning of the vertebrate body plan. Developmental Biology, 2010, 344(1): 7-15 ( ).
- DOI: 10.1016/j.ydbio.2010.04.024
- Yurchenko A.A., Deniskova T.E., Yudin N.S., Dotsev A.V., Khamiruev T.N., Selionova M.I., Egorov S.V., Reyer H., Wimmers K., Brem G., Zinovieva N.A., Larkin D.M. High-density genotyping reveals signatures of selection related to acclimation and economically important traits in 15 local sheep breeds from Russia. BMC Genomics, 2019, 20(Suppl 3): 294 ( ).
- DOI: 10.1186/s12864-019-5537-0
- Ahbara A., Bahbahani H., Almathen F., Al Abri M., Agoub M.O., Abeba A., Kebede A., Musa H.H., Mastrangelo S., Pilla F., Ciani E., Hanotte O., Mwacharo J.M. Genome-wide variation and putative candidate regions and genes associated with fat deposition and tail morphology in Ethiopian indigenous sheep. Front. Genet., 2019, 9: 699 ( ).
- DOI: 10.3389/fgene.2018.00699
- Ahbara A., Musa H., Clark E., Robert C., Watson M., Abeba A., Latairish S., Hanotte O., Mwacharo J. OP196 indigenous African sheep genomes reveal insights on fat-tail deposition and morphology. Proc. 37th International Conference on Animal Genetics. Lleida, Spain, 2019: 56.
- Zhi D., Da L., Liu M., Cheng C., Zhang Y., Wang X., Li X., Tian Z., Yang Y., He T., Long X., Wei W., Cao G. Whole genome sequencing of Hulunbuir short-tailed sheep for identifying candidate genes related to the short-tail phenotype. G3: Genes, Genomes, Genetics, 2018, 8(2): 377-383 ( ).
- DOI: 10.1534/g3.117.300307
- Liu Z., Wang H., Liu R., Mingming W.U., Zhang S., Zhang L., Zhao F., Du L., Wei C. Genome-wide detection of selection signatures of distinct tail types in sheep popula-tions. Acta Veterinaria et Zootechnica Sinica, 2015, 46(10): 1721-1732 ( ).
- DOI: 10.11843/j.issn.0366-6964.2015.10.004
- Mwacharo J.M., Kim E.S., Elbeltagy A.R., Aboul-Naga A.M., Rischkowsky B.A., Rothschild M.F. Genomic footprints of dryland stress adaptation in Egyptian fat-tail sheep and their divergence from East African and western Asia cohorts. Sci. Rep., 2017, 7(1): 17647 ( ).
- DOI: 10.1038/s41598-017-17775-3
- Сермягин А.А., Гладырь Е.А., Харитонов С.Н., Ермилов А.Н., Стрекозов Н.И., Брем Г., Зиновьева Н.А. Полногеномный анализ ассоциаций с продуктивными и репродуктивными признаками у молочного скота в российской популяции голштинской породы. Сельскохозяйственная биология, 2015, 51(2): 182-193 ( ).
- DOI: 10.15389/agrobiology.2016.2.182rus
- Jonas E., Thomson P.C., Raadsma H.W. Genome-wide association study and fine mapping of QTL on OAR 21 for body weight in sheep. Proc. 9th World Congress on Genetics Applied to Livestock Production. Leipzig, 2010.
- Kominakis A., Hager-Theodorides A.L., Zoidis E., Saridaki A., Antonakos G., Tsiamis G. Combined GWAS and 'guilt by association'-based prioritization analysis identifies functional candidate genes for body size in sheep. Genet. Sel. Evol., 2017, 49: 41 ( ).
- DOI: 10.1186/s12711-017-0316-3
- Xu S.S., Ren X., Yang G.L., Xie X.L., Zhao Y.X., Zhang M., Shen Z.Q., Ren Y.L., Gao L., Shen M., Kantanen J., Li M.H. Genome-wide association analysis identifies the genetic basis of fat deposition in the tails of sheep (Ovis aries). Anim. Genet., 2017, 48(5): 560-569 ( ).
- DOI: 10.1111/age.12572
- Bhanuprakash V., Chhotaray S., Pruthviraj D.R., Rawat C., Karthikeyan A., Panigrahi M. Copy number variation in livestock: a mini review. Veterinary World, 2018, 11(4): 535-541 ( ).
- DOI: 10.14202/vetworld.2018.535-541
- Stranger B.E., Forrest M.S., Dunning M., Ingle C.E., Beazley C., Thorne N., Redon R., Bird C.P., de Grassi A., Lee C., Tyler-Smith C., Carter N., Scherer S.W., Tavaré S., Deloukas P., Hurles M.E., Dermitzakis E.T. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science, 2007, 315(5813): 848-853 ( ).
- DOI: 10.1126/science.1136678
- Liu G.E., Ventura M., Cellamare A., Chen L., Cheng Z., Zhu B., Li C., Song J., Eichler E.E. Analysis of recent segmental duplications in the bovine genome. BMC Genomics, 2009, 10: 571 ( ).
- DOI: 10.1186/1471-2164-10-571
- Zhang F., Gu W., Hurles M.E., Lupski J.R. Copy number variation in human health, disease, and evolution. Annual Review of Genomics and Human Genetics, 2009, 10: 451-481 ( ).
- DOI: 10.1146/annurev.genom.9.081307.164217
- Fontanesi L., Beretti F., Martelli P.L., Colombo M., Dall'Olio S., Occidente M., Portolano B., Casadio R., Matassino D., Russo V. A first comparative map of copy number variations in the sheep genome. Genomics, 2011, 97(3): 158-165 ( ).
- DOI: 10.1016/j.ygeno.2010.11.005
- Jenkins G.M., Goddard M.E., Black M.A., Brauning R., Auvray B., Dodds K.G., Kijas J.W., Cockett N., McEwan J.C. Copy number variants in the sheep genome detected using multiple approaches. BMC Genomics, 2016, 17: 441 ( ).
- DOI: 10.1186/s12864-016-2754-7
- Liu J., Zhang L., Xu L., Ren H., Lu J., Zhang X., Zhang S., Zhou X., Wei C., Zhao F., Du L. Analysis of copy number variations in the sheep genome using 50K SNP BeadChip array. BMC Genomics, 2013, 14: 229 ( ).
- DOI: 10.1186/1471-2164-14-229
- Ma Y., Zhang Q., Lu Z., Zhao X., Zhang Y. Analysis of copy number variations by SNP50 BeadChip array in Chinese sheep. Genomics, 2015, 106(5): 295-300 ( ).
- DOI: 10.1016/j.ygeno.2015.08.001
- Zhu C., Fan H., Yuan Z., Hu S., Ma X., Xuan J., Wang H., Zhang L., Wei C., Zhang Q., Zhao F., Du L. Genome wide detection of CNVs in Chinese indigenous sheep with different types of tails using ovine high-density 600K SNP arrays. Scientific Reports, 2016, 6: 27822 ( ).
- DOI: 10.1038/srep27822
- Ma Q., Liu X., Pan J., Ma L., Ma Y., He X., Zhao Q., Pu Y., Li Y., Jiang L. Genome-wide detection of copy number variation in Chinese indigenous sheep using an ovine high-density 600 K SNP array. Scientific Reports, 2017, 7: 912 ( ).
- DOI: 10.1038/s41598-017-00847-9
- Hoeijmakers W.A., Bártfai R., Stunnenberg H.G. Transcriptome analysis using RNA-Seq. In: Malaria. Methods in molecular biology (methods and protocols) /R. Ménard (ed.). Humana Press, Totowa, NJ, 2012, vol. 923: 221-239 ( ).
- DOI: 10.1007/978-1-62703-026-7_15
- Shendure J. The beginning of the end for microarrays? Nature Methods, 2008, 5(7): 585-587 ( ).
- DOI: 10.1038/nmeth0708-585
- Oshlack A., Robinson M.D., Young M.D. From RNA-seq reads to differential expression results. Genome Biol., 2010, 11(12): 220 ( ).
- DOI: 10.1186/gb-2010-11-12-220
- Wang X., Zhou G., Xu X., Geng R., Zhou J., Yang Y., Yang Z., Chen Y. Transcriptome profile analysis of adipose tissues from fat and short-tailed sheep. Gene, 2014, 549(2): 252-257 ( ).
- DOI: 10.1016/j.gene.2014.07.072
- Kang D., Zhou G., Zhou S., Zeng J., Wang X., Jiang Y., Yang Y., Chen Y. Comparative transcriptome analysis reveals potentially novel roles of Homeobox genes in adipose deposition in fat-tailed sheep. Sci. Rep., 2017, 7(1): 14491 ( ).
- DOI: 10.1038/s41598-017-14967-9
- Ma L., Li Z., Cai Y., Xu H., Yang R., Lan X. Genetic variants in fat- and short-tailed sheep from high-throughput RNA-sequencing data. Anim. Genet., 2018, 49(5): 483-487 ( ).
- DOI: 10.1111/age.12699
- Ma L., Zhang M., Jin Y., Erdenee S., Hu L., Chen H., Cai Y., Lan X. Comparative transcriptome profiling of mRNA and lncRNA related to tail adipose tissues of sheep. Front. Genet., 2018, 9: 365 ( ).
- DOI: 10.3389/fgene.2018.00365
- Li B., Qiao L., An L., Wang W., Liu J., Ren Y., Pan Y., Jing J., Liu W. Transcriptome analysis of adipose tissues from two fat-tailed sheep breeds reveals key genes involved in fat deposition. BMC Genomics, 2018, 19(1): 338 ( ).
- DOI: 10.1186/s12864-018-4747-1
- Avilés C., Polvillo O., Peña F., Juárez M., Martínez A.L., Molina A. Associations between DGAT1, FABP4, LEP, RORC, and SCD1 gene polymorphisms and fat deposition in Spanish commercial beef. Journal of Animal Science, 2013, 91(10): 4571-4577 ( ).
- DOI: 10.2527/jas.2013-6402
- Xu Q.L., Tang G.W., Zhang Q.L., Huang Y.K., Liu Y.X., Quan K., Zhu K.Y., Zhang C.X. The FABP4 gene polymorphism is associated with meat tenderness in three Chinese native sheep breeds. Czech Journal of Animal Science, 2011, 56(1): 1-6.
- Bokor S., Legry V., Meirhaeghe A., Ruiz J.R., Mauro B., Widhalm K., Manios Y., Amouyel P., Moreno L.A., Molnàr D., Dallongeville J., HELENA Study group. Single-nucleotide polymorphism of CD36 locus and obesity in European adolescents. obesity, 2010, 18(7): 1398-1403 ( ).
- DOI: 10.1038/oby.2009.412
- Kern P.A., Di Gregorio G.B., Lu T., Rassouli N., Ranganathan G. Adiponectin expression from human adipose tissue: relation to obesity, insulin resistance, and tumor necrosis factor-alpha expression. Diabetes, 2003, 52(7): 1779-1785 ( ).
- DOI: 10.2337/diabetes.52.7.1779
- Zhang L., Yang M., Li C., Xu Y., Sun J., Lei C., Lan X., Zhang C., Chen H. Identification and genetic effect of a variable duplication in the promoter region of the cattle ADIPOQ gene. Anim. Genet., 2014, 45(2): 171-179 ( ).
- DOI: 10.1111/age.12112
- Erdenee S., Li J., Kang Z., Xu H., Zang R., Cao X., Yang J., Cai Y., Lan X. Sheep zinc finger proteins 395 (ZNF395): insertion/deletion variations, associations with growth traits, and mRNA expression. Animal Biotechnology, 2019, 19: 1-8 ( ).
- DOI: 10.1080/10495398.2019.1585865
- Bakhtiarizadeh M.R., Moradi-Shahrbabak M., Ebrahimie E. Underlying functional genomics of fat deposition in adipose tissue. Gene, 2013, 521(1): 122-128 ( ).
- DOI: 10.1016/j.gene.2013.03.045
- Ruixia X., Lei G., Weili Z., Wei Z., Guangchao S., Shangquan G., Guoqing S. [Analysis of FABP4 expression pattern in rump fat deposition and metabolism of Altay sheep]. Yi Chuan, 2015, 37(2): 174-182 ( ).
- DOI: 10.16288/j.yczz.14-369
- Bakhtiarizadeh M.R., Salami S.A. Identification and expression analysis of long noncoding RNAs in fat-tail of sheep breeds. G3: Genes, Genomes, Genetics, 2019, 9(4): 1263-1276 ( ).
- DOI: 10.1534/g3.118.201014
- Jiao X.L., Jing J.J., Qiao L.Y., Liu J.H., Li L.A., Zhang J., Jia X.L., Liu W.Z. Ontogenetic expression of Lpin2 and Lpin3 genes and their associations with traits in two breeds of Chinese fat-tailed sheep. Asian-Australasian Journal of Animal Sciences, 2016, 29(3): 333-342 ( ).
- DOI: 10.5713/ajas.15.0467
- Zakariapour Bahnamiri H., Ganjkhanlou M., Zali A., Sadeghi M., Moradi Shahrbabak H., Nehzati Paghaleh G.A. Expression of genes related to liver fatty acid metabolism in fat-tailed and thin-tailed lambs during negative and positive energy balances. J. Anim. Physiol. Anim. Nutr., 2019, 103(2): 427-435 ( ).
- DOI: 10.1111/jpn.13036
- Zeng J., Zhou S.W., Zhao J., Jin M.H., Kang D.J., Yang Y.X., Wang X.L., Chen Y.L. Role of OXCT1 in ovine adipose and preadipocyte differentiation. Biochemical and Biophysical Research Communications, 2019, 512(4): 779-785 ( ).
- DOI: 10.1016/j.bbrc.2019.03.128
- Ledur M.C., Navarro N., Pérez-Enciso M. Large-scale SNP genotyping in crosses between outbred lines: how useful is it? Heredity, 2009, 105(2): 173-182 ( ).
- DOI: 10.1038/hdy.2009.149


