Dimensionality Reduction using Genetic Algorithm for Improving Accuracy in Medical Diagnosis
Автор: D. Asir Antony Gnana Singh, E. Jebamalar Leavline, R. Priyanka, P. Padma Priya
Журнал: International Journal of Intelligent Systems and Applications(IJISA) @ijisa
Статья в выпуске: 1 vol.8, 2016 года.
Бесплатный доступ
The technological growth generates the massive data in all the fields. Classifying these high-dimensional data is a challenging task among the researchers. The high-dimensionality is reduced by a technique is known as attribute reduction or feature selection. This paper proposes a genetic algorithm (GA)-based features selection to improve the accuracy of medical data classification. The main purpose of the proposed method is to select the significant feature subset which gives the higher classification accuracy with the different classifiers. The proposed genetic algorithm-based feature selection removes the irrelevant features and selects the relevant features from original dataset in order to improve the performance of the classifiers in terms of time to build the model, reduced dimension and increased accuracy. The proposed method is implemented using MATLAB and tested using the medical dataset with various classifiers namely Naïve Bayes, J48, and k-NN and it is evident that the proposed method outperforms other methods compared.
Attribute reduction, Naive Bayes classifier, genetic algorithm
Короткий адрес: https://sciup.org/15010788
IDR: 15010788
Текст научной статьи Dimensionality Reduction using Genetic Algorithm for Improving Accuracy in Medical Diagnosis
Published Online January 2016 in MECS
Data mining is a process of extracting the interesting patterns or information from the available data to extract the knowledge. The data mining plays a vital role in knowledge discovery process. It generates the pattern in terms of classification models to predict the unknown data from known data by this way the essential knowledge is obtained from the huge volumes of data. The learning algorithms which are used to build the classification model are formally categorized into two types: supervised and unsupervised learning algorithm.
The supervised learning algorithm deals with the labeled data for developing the models known as classifiers. The unsupervised learning algorithm learns the unlabeled data and develops the model for prediction. A typical example for this case is clustering. The supervised learning algorithm is commonly adopted in the feature selection algorithms for evaluating the quality of the feature subsets. In some cases, the supervised learning algorithm is used to evaluate the performance of the feature selection algorithms in terms of classification accuracy.
Feature selection is a process of selecting the relevant attributes and removing the irrelevant and redundant attributes. Feature selection provides three main benefits while constructing predictive models: improves the model interpretability, shortens the training times, and enhanced generalization by reducing over fitting. Further, the feature selection algorithm is classified into three types namely wrapper, filter, and embedded method. The wrapper method uses a supervised learning algorithm to evaluate the feature to select the significant feature subsets from a dataset.
The embedded method uses the supervised learning algorithm as a part of the feature selection process. The filter method uses the any one of the statistical or mathematical measures to select the significant feature from the dataset without help of the supervised learning algorithm. Each classifier follows its own learning method. In general the data preprocessing can improve the performance of the classification algorithm and decrease the computational complexity.
GA has been known to be very adaptive and efficient method of feature selection. It is basically a searching algorithm based on natural genetics and natural selection. It is an optimization technique, a population-based and algorithmic search heuristic method. The operations in a GA are iterative procedures manipulating one population of chromosomes to produce a new population through genetic functions such as crossover (recombination between two single chromosomes) and mutation (randomly changes the chromosomes).
This paper proposes a wrapper-based feature selection method combining the supervised learning algorithm for evaluate the feature subset searched by the genetic algorithm from a high-dimensional data. In order to evaluate the performance of the proposed method the classifiers namely k- Nearest Neighbor Algorithm (k-NN), Naïve Bayes (NB), and J48 are used.
The rest of the paper is organized as follows: Section II reviews the literature. In Section III the proposed method is described. In Section IV, the implementation details are elaborated. Section V presents and discusses the experimental results and Section VI concludes the paper.
-
II. Related Works
This section details the various research works which are related to the proposed method. Lior Rokach et al. developed a genetic algorithm-based feature selection to solve the classification problem. In this paper, a new encoding algorithm was also proposed for evaluating the fitness function of multiple, obvious tree classifier [1]. Magnus Erik et al. presented a genetic algorithm for feature selection in data mining and they showed that the feature selection can be used to avoid feature induced over fitting [2].
M. Analoui et al. proposed a feature reduction of nearest neighbor classifier using genetic algorithm. In this approach, each value is first normalized by a linear equation then scaled by the weight prior to testing, training and classification [3]. The Pier luca lanzi et al. developed a fast feature selection with genetic algorithm. In this paper, a filter approach is used and also this approach shows that the feature selection requires a large amountof CPU time to reach a good solution on large datasets [4].
The Zili Zhang and Pengyi Yang proposed an ensemble of classifiers with GA-based feature selection. The authors of this paper developed a novel hybrid algorithm which is combination of multi objective genetic algorithm and ensemble of classifiers. The GA-ensemble approach was evaluated on various datasets and compared its performance using various classifiers [5].
Li Zhuo et al. presented a GA-based wrapper feature selection method for classification of hyper spectral images using support vector machine. This method follows wrapper and filter approach [6]. Amira Sayed et al. presented a genetic algorithm-based feature selection method for anomaly detection. In this paper, several feature selection techniques were used including principle component analysis (PCA), sequential floating, and correlation-based feature selection [7].
Laetitia Jourdan developed a GA for feature selection in data mining for genetics. They specified two approaches: first one is heuristic approach and second one is clustering-based feature selection approach. Diseases dataset such as obesity, diabetes were used to test their method [8]. Younes Chtioui et al. presented a feature selection by genetic algorithm which is an application to seed discrimination by artificial vision. The performance of this method was evaluated on a practical pattern recognition problem, which deals with the discrimination between four seeds (two cultivated and two adventitious seed species). In this paper, nearest neighbor (k-NN) classification method was used for classification [9].
Cheng-Lung Huang et al. proposed a feature selection based on the GA and optimization with SVM. The ultimate aim was to optimize the feature subset and parameters without deteriorating the SVM classification accuracy. In this paper GA was compared with the grid algorithm [10]. The Yi Sun and Lijun Yin presented a feature selection based on the genetic algorithm to solve the problem of face recognition and a generic model is used to construct the features for 3D facial expressions. This approach evidences that this type of feature selection is very useful to face modeling [11].
Haleh Vafaie et al. presented a feature selection approach based on greedy-like search and genetic algorithm. In this paper, the authors presented a comparison between these two approaches, and identified that the GA-based method produced the better performance [12]. A. Srikrishna et al. also presented GAbased feature selection. This method reduces the computational complexity and achieves better performing clustering [13]. Haleh Vafaie et al. presented a genetic algorithm-based approach to develop the rules for identifying the suitable variable subset for the texture classification tasks and observed that it also can be used in rule induction machine learning systems [14]. W. Siedlecki et al. presented the usage of the genetic algorithm for selecting the necessary feature subsets form the large dataset [15].
This genetic algorithm can also be employed for various applications in machine learning. It enhances the object grouping techniques such as clustering in unsupervised learning to obtain the optimal clustercenters for object clustering [16]. In pattern recognition, the genetic algorithm can be used for detection operations including face recognition [17], handwritten digits [18], gas-insulated system and etc. [19]. Further, the GA based approaches are employed to search the needed or unneeded image region for split or merging process in image segmentation [20]. This is also used as a searching mechanism in flow shop sequencing [21].
From this literature review it is obvious that the GAbased searching technique plays a significant role in the field of the data mining as well as the feature selection process. Hence, genetic algorithm is employed in our proposed approach as the searching algorithm in the feature selection process. The supervised learning algorithm is employed for evaluating the feature subsets hence the proposed approach is a wrapper-based feature selection method.
-
III. Proposed Genetic Algorithm Based Feature Selection
GA was originally used to select binary strings and a number of authors have discussed the use of GA in feature selection. One of the significant characteristics of GA is that it has been evolved in such a way to explore inter-dependencies between the bits in a string and hence it is very much suitable for the feature selection problem where we look for dependencies between features and select the best ones that contribute to the classification or recognition performance. In most of the cases it performs better than the forward and backward search algorithms in terms of number of evolutions to reach minimum [22].
The probability of getting an optimal feature subset for classification is high when GA is employed for feature selection with suitable fitness functions and possible considerations [23]. Taking these advantages, a feature selection scheme based on genetic algorithm is proposed. The proposed wrapper based feature selection method takes the advantage of the supervised learning algorithm to evaluate the significance of the feature subset and employs the genetic algorithm to optimize the searching of features in feature selection process. The performance of the proposed method is tested on the medical dataset with various classifiers.
-
A. Proposed Feature Selection Algorithm
Genetic algorithm (GA) is a method for moving to a new population from an existing population of chromosomes using a natural selection method. It has two operators namely crossover and mutation. Crossover exchanges subparts of two chromosomes or it performs recombination between two single chromosomes. Mutation randomly changes the allele values of some locations in the chromosome. GA evaluates the fitness of each and every individual; this means that the superiority of the results is achieved through a fitness function. The suitable chromosome has higher probability to choose for the next generation formation.
If the fittest of the chromosome in a population cannot meet the requirement, crossover and mutation functions will be carried out. The functions are carried out repeatedly until the acceptable result is obtained. The operations of the algorithm are explained in the following section and Table 1 shows the terminology of human genetics and their equivalent in GA.
-
B. Operations of the Proposed Algorithm
The operational steps of the proposed algorithm are population initialization, performing crossover and mutation, fitness evaluation, and stopping criteria as depicted in Figure 1.
This algorithm initially assigns binary search space, as the chromosomes are bit strings and each bit represents each feature of the training dataset. This initial population is created (randomly) with the assumption that a gene value ‘1’ represents the particular feature that is to be selected for evaluation which maintains the same position of the gene value (same index) and if it is ‘0’, the feature is not selected which maintains the position of the gene value.
The parents are randomly selected and their length is same as the total number of features (i.e. if the total number of feature is 5 then the chromosomes contain 5 genes that means 5 bits).
Second step performs the crossover operation. Two parents i.e. two individuals (chromosomes) are combined together to form a child (new chromosome). This crossover operation is carried out with four methods based on the logical operations such as single point crossover operation, XOR operation, OR operation, and AND operation the sample of these operations are depicted in Figure 3 where P1 and P2 are parents, and the C 1 and C 2 are the children.
Table 1. Terminology of human genetics and their equivalent in GA
|
S. No. |
Human genetics terminology |
GA terminology |
|
1 |
Chromosomes |
Bit strings |
|
2 |
Genes |
Features |
|
3 |
Allele |
Feature value |
|
4 |
Locus |
Bit position |
|
5 |
Genotype |
Encoded string |
|
6 |
Phenotype |
Decoded genotype |
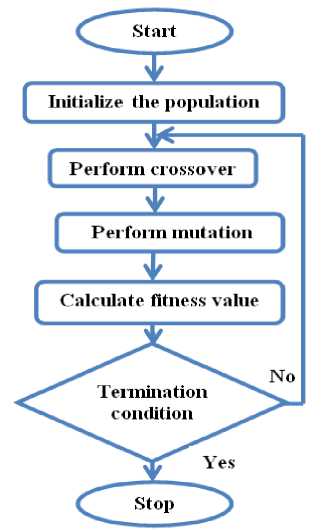
Fig.1. Flowchart representation of the proposed algorithm
Third step performs the mutation operation that randomly changes the allele values from crossover child (C) to produce the next generation. Figure 4 shows the flip bit-based crossover on a child (C) with the random mute at the fourth position of C (bit ‘1’). The produced next generation (new chromosome : 1 1 1 0 0 1 1 1) denotes that as mentioned earlier the dataset contains the total number of 8 features since the chromosome contains 8 bits and each bit represents the index of each feature of the dataset. Then the features corresponding to the 0 chromosome value are discarded thereby the new subset is obtained from the dataset such as f1,f2,f3,f6,f7,f8 where the f4 and f5 are discarded since the bit position representing 0.
|
Chromosome |
1 |
0 |
1 |
1 |
0 |
0 |
0 |
1 |
Fig.2. Representation of the chromosome for the dataset containing 8 features
|
Pi: |
1 |
0 |
1 |
0 |
0 |
1 |
1 |
1 |
|
P2: |
1 |
1 |
1 |
1 |
0 |
1 |
1 |
0 |
|
Ci: |
1 |
0 |
1 |
0 |
0 |
0 |
1 |
0 |
|
C2: |
1 |
1 |
1 |
1 |
0 |
1 |
1 |
1 |
(a)
|
Pi: |
1 |
0 |
1 |
0 |
0 |
1 |
1 |
1 |
|
P;: |
1 |
1 |
1 |
1 |
0 |
0 |
1 |
0 |
|
C: |
0 |
1 |
0 |
1 |
0 |
1 |
0 |
1 |
(b)
|
Pi: |
1 |
0 |
1 |
0 |
0 |
1 |
1 |
1 |
|
P2: |
1 |
1 |
1 |
1 |
0 |
0 |
1 |
0 |
|
C: |
1 |
1 |
1 |
1 |
0 |
1 |
1 |
1 |
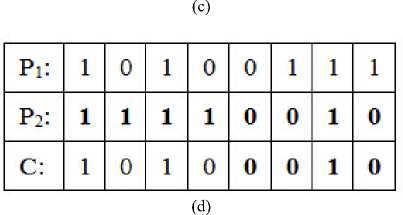
Fig.3. The crossover operation (a) single point (b) XOR (c) OR (d) AND
In the fourth step, the fitness value is calculated from the obtained feature subset from step 3. The classification accuracy is considered as the fitness value to validate the generated subset. Therefore the NB classifier is used to calculate the classification accuracy of the generated subset. If the classification accuracy (fitness value) satisfies the termination condition, this feature subset is considered as the selected feature subset otherwise step 2 to step 4 are followed in an iterative manner until the termination condition is satisfy.
-
IV. Implementation
-
A. System Specification and Dataset
The proposed method is implemented using MATLAB with the system specification 4 GB RAM, 120 GB Hard Disk, Windows Vista operating system, Dual core Intel Processor. The performance of the proposed method is evaluated using the diabetes dataset which contains the 8 attributes and 768 instances.
-
B. Performance Evaluation of the Proposed Method
Formally, the classification algorithms are used to evaluate the significance of the feature subset selected by the feature selection algorithm. The classification algorithms are a type of supervised machine learning. In this paper, the classification algorithms namely Naïve Bayes (NB), J48 and IB1 are used to evaluate the performance of the feature selection methods.
-
1) NB Classification Algorithm
This is a probabilistic based algorithm that builds a probabilistic model based on the dataset with the selected feature. This probabilistic model is known as classifier. The input is given in the form of instances. The instance can be denoted as X=(x 1 , x 2 , …, x n ), where x i denotes the feature values on ‘n’ number of features that are selected by the feature selection algorithm. The probability of predicting the possible class label c i (i = 1,2,…, k) for a particular instance xi is denoted as p(ci| x1,x2,…,xn). In this context, the posterior and prior probability are calculated in order to predict the class label ci for a given instance xi [24,25].
-
2) J48 Classification Algorithm
This is a tree based classification algorithm. Initially, it receives the dataset and calculates the weight of the features using any feature ranking algorithm such as information gain. The node that contains higher weight than that node is under consideration as the root node to form a tree structure from the training dataset. Then the tree is further expanded through the higher weight feature node until all the nodes contain the single class label. The prediction of the class label for a given instance is carried out based on the constructed tree. This tree is also called as decision tree for predicting a label of a particular instance [26].
-
3) IB1 Classification Algorithm
This is an instance based classification algorithm. In this algorithm, the nearest neighbor principle is followed in order to carry out the prediction of the label for an unlabeled instance [27].
-
C. Performance Evaluation Metrics
The performance is evaluated in terms of runtime, number of feature selected [28], and the accuracy produced with the various classifiers such as Naïve Bayes, k-NN, and J48
-
1) Classification Accuracy
The classification accuracy is calculated using the formula A=(a+d)/(a+b+c+d) where ‘a’ is the number of correctly classified negative instances, ‘b’ is the number of incorrectly classified positive instances, ‘c’ is the number of incorrectly classified negative instances, and ‘d’ is the number of correctly classified positive instances.
-
2) Number of Features Selected
This is the number of features selected after applying the proposed GA-based feature selection algorithm.
-
3) Runtime
This is the total runtime of the proposed algorithm in MATLAB environment with the specifications mentioned earlier and it is measured in seconds.
-
D. Experiment Procedure
The experimental procedure is illustrated in Figure 5. Initially, the original dataset with full features is given to the proposed algorithm and then the original dataset is reduced to a set of selected features. Then these selected features are divided into two dataset namely training dataset and test dataset. Then the NB, k-NN, and J48 supervised learning algorithms are used to build the classifier using the training dataset and the test dataset is used to evaluate the accuracy of the classifier in the iterative manner (i.e. 10-fold cross validation) and averaged to obtain the accuracy.
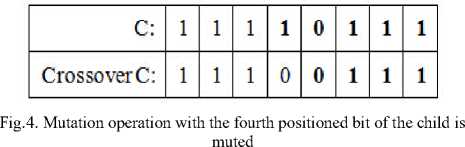
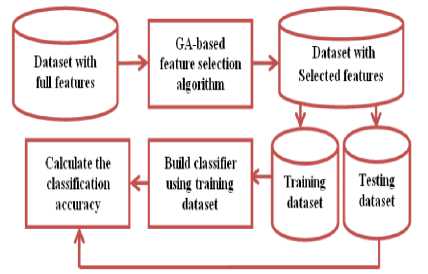
Fig.5. Experimental procedure
The experiment is conducted with four different methods as illustrated in Figure 5. In order to conduct the experiment and obtain the results for the proposed algorithm, the four experimental methods are formed namely M1, M2, M3, and M4by changing the operations in crossover and mutation steps of the proposed algorithm as shown in Table 2.
-
V. Results and Discussion
The obtained results of classification accuracy, number of feature selected, and the runtime of the different methods are tabulated in Table 2. The experimental methods M1, M2, M3, and M4 differ from each other in terms of crossover operation. M1 uses AND operation, M2 uses XOR operation, M3 employs OR operation and M4 uses a single point crossover operation. For all the four methods, flip bit method is used to perform mutation operation.
Figure 6 to Figure 8 show the results on number of feature selected, classification accuracy, and runtime of the different methods. From Figure 6, it is evident that the methods M1 and M2 are reducing the number of features in similar quantities. M3 and M4 are performing almost equally. The methods M1 and M2 have reduced more number of features than M3 and M4.
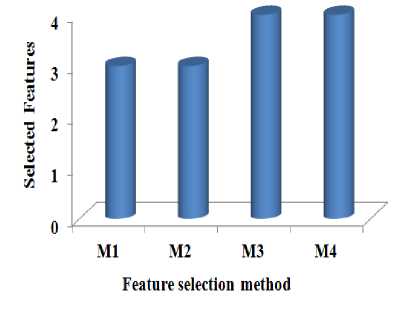
Fig.6. The number of feature selected with respect to the feature selection method
Ml М2 М3 M4
Feature selection method
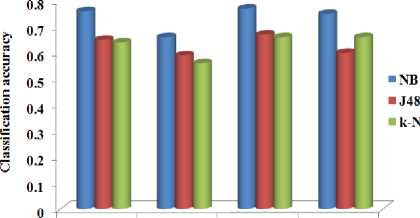
Fig.7. The classification accuracy against the feature selection method
Figure 7 shows that the NB classifier produces the better accuracy with all the methods compared to other classifiers. J48 produces better accuracy compared to the k-NN classifier except with the method M4. The method M3 produces the overall better accuracy compared to all other methods with all the classifiers.
2.5
Ml М2 М3 M4
Feature selection method
Table 2. Comparison of number of feature selected, runtime, and classification accuracy against the four feature selection methods
|
Proposed algorithm |
Classification accuracy |
|||||||
|
Experimental Method |
Crossover operation |
Mutation operation |
No. of selected features |
No. of Iterations |
Runtime (Sec) |
NB |
J48 |
k-NN |
|
M1 |
AND |
Flip bit |
3 |
10 |
1.71 |
0.76 |
0.65 |
0.64 |
|
M2 |
XOR |
Flip bit |
3 |
10 |
2.10 |
0.66 |
0.59 |
0.56 |
|
M3 |
OR |
Flip bit |
4 |
10 |
2.10 |
0.77 |
0.67 |
0.66 |
|
M4 |
Single point |
Flip bit |
4 |
10 |
1.90 |
0.75 |
0.60 |
0.66 |
-
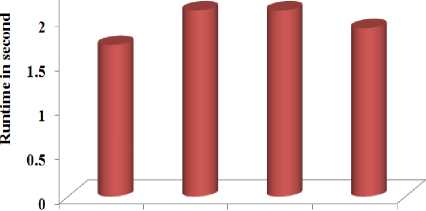
Fig.8. The runtime against the feature selection method
From Figure 8, it is observed that the method M1 takes less time compared to all other methods and methods M2 and M3 consume more or less similar time.
In general, the wrapper approach produces the higher accuracy for the supervised learner which is adopted for evaluating the feature subset thus the proposed system produces the better accuracy with NB classifier.
-
VI. Conclusion and Future Enhancement
This paper proposed a genetic algorithm-based wrapper feature selection for medical data classification. The experiment is conducted with four experimental strategies. The proposed system accommodates the genetic algorithm (GA) to search and form the feature subsets and the Naïve Bayes classifier as the evaluation tool to select the significant feature subset. The performance of the proposed method is evaluated with the well established classifiers such as Naïve Bayes, J48 and k-NN. The performance of the proposed method is analyzed in terms of number of features reduced, algorithm runtime, and the classification accuracy produced with different classifiers. From the conducted experiments, it is evident that the proposed method outperforms other methods compared and produces better accuracy for the classifier which is adopted as the feature evaluation mechanism in the feature selection process. Hence this proposed method is well suited for the medical application where the classification algorithm is predefined.
As a future enhancement of this proposed method, this method can be combined with other natural selection algorithms for searching strategy and other classifiers as evaluation mechanism.
Список литературы Dimensionality Reduction using Genetic Algorithm for Improving Accuracy in Medical Diagnosis
- Rokach Lior, "Genetic algorithm-based feature set partitioning for classification problems," Pattern Recogn, vol. 41, pp.1676-1700, 2008.
- Magnus Erik and Hvass Pedersen, Genetic Algorithms for Feature Selection in Data Mining, Pedersen (971055) Daimi, University of Aarhus, November 2003.
- Analoui, M., and M. Fadavi Amiri. "Feature reduction of nearest neighbor classifiers using genetic algorithm." World Acad Sci Eng Technol, vol. 17, pp. 36-39, 2003.
- Lanzi, Pier Luca, "Fast feature selection with genetic algorithms: a filter approach," IEEE International Conference on Evolutionary Computation, pp. 537-540, 1997.
- Zhang, Zili, and Pengyi Yang, "An ensemble of classifiers with genetic algorithm Based Feature Selection," IEEE intelligent informatics bulletin, vol. 9, pp. 18-24, 2008.
- Zhuo, Li, Jing Zheng, Xia Li, Fang Wang, Bin Ai, and Junping Qian. "A genetic algorithm based wrapper feature selection method for classification of hyperspectral images using support vector machine," Geoinformatics 2008 and Joint Conference on GIS and Built Environment: Classification of Remote Sensing Images, pp. 71471J-71471J, 2008.
- Aziz, Amira Sayed A., Ahmad Taher Azar, Mostafa A. Salama, Aboul Ella Hassanien, and SE-O. Hanafy, "Genetic algorithm with different feature selection techniques for anomaly detectors generation." IEEE Federated Conference on Computer Science and Information Systems (FedCSIS), pp. 769-774, 2013.
- Jourdan, Laetitia, Clarisse Dhaenens, and El-Ghazali Talbi, "A genetic algorithm for feature selection in data-mining for genetics." 4th Metaheuristics International Conference Porto, pp. 29-34, 2001.
- Chtioui, Younes, Dominique Bertrand, and Dominique Barba, "Feature selection by a genetic algorithm. Application to seed discrimination by artificial vision." J Sci Food Agric, vol. 76, pp. 77-86, 1998.
- Huang, Cheng-Lung, and Chieh-Jen Wang, "A GA-based feature selection and parameters optimization for support vector machines", Expert Syst Appl, vol. 31, pp. 231-240, 2006.
- Sun, Yi, and Lijun Yin. "A genetic algorithm based feature selection approach for 3D face recognition." The Biometric Consortium Conference, (Hyatt Regency Crystal City, Arlington, Virginia USA), 2005.
- Vafaie, Haleh, and Ibrahim F. Imam, "Feature selection methods: genetic algorithms vs. reedy-like search," International Conference on Fuzzy and Intelligent Control Systems, 1994.
- Srikrishna, A., B. Eswara Reddy, and V. Sesha Srinivas. "Automatic Feature Subset Selection using Genetic Algorithm for Clustering." Int J Recent Trends Eng Tech, vol. 9, pp. 85-89, 2013.
- Vafaie, Haleh, and Kenneth De Jong. "Genetic algorithms as a tool for feature selection in machine learning." Fourth IEEE International Conference on Tools with Artificial Intelligence, Arlington, VA, pp. 200 – 203, 1992.
- Maulik, Ujjwal, and Sanghamitra Bandyopadhyay. "Genetic algorithm-based clustering technique." Pattern Recogn, vol. 33, pp. 1455-1465, 2000.
- Siedlecki, Wojciech, and Jack Sklansky. "A note on genetic algorithms for large-scale feature selection." Pattern Recogn Lett, vol. 10, pp. 335-347, 1989.
- Sarawat Anam, Md. Shohidul Islam, M.A. Kashem, M.N. Islam, M.R. Islam and M.S. Islaml. "Face recognition using genetic algorithm and back propagation neural network." International MultiConference of Engineers and Computer Scientists. 2009.
- Cho, Sung-Bae. "Pattern recognition with neural networks combined by genetic algorithm," Fuzzy Set Syst, vol. 103, pp. 339-347, 1999.
- Ziomek, W., M. Reformat and E. Kuffel. "Application of genetic algorithms to pattern recognition of defects in GIS," IEEE T Dielect El In, vol. 7, pp. 161-168, 2000.
- Chun, Dae N. and Hyun S. Yang. "Robust image segmentation using genetic algorithm with a fuzzy measure," Pattern Recogn, vol. 29, pp. 1195-1211, 1996.
- Reeves, CR. "A genetic algorithm for flowshop sequencing." Comput Oper Res, vol. 22, pp. 5-13, 1995.
- Hunter, A. “Feature selection using probabilistic neural networks,” Neural Comput Appl, vol. 9, pp. 124-132, 2000.
- Rubiyah Yusof, Uswah Khairuddin and Marzuki Khalid. “A New Mutation Operation for Faster Convergence in Genetic Algorithm Feature Selection,” IJICIC, vol. 8, pp. 7363-7378, 2012.
- Martinez-Arroyo, M. and Sucar, LE. “Learning an optimal naive bayes classifier”, 18th IEEE International Conference on Pattern Recognition, vol. 3, pp. 1236-1239, 2006.
- Zia, T., Abbas, Q., and Akhtar, MP. “Evaluation of Feature Selection Approaches for Urdu Text Categorization,” I.J. Intelligent Systems and Applications, vol. 7, pp. 33-40, 2015.
- Kotsiantis, SB. “Supervised Machine Learning: A Review of Classification Techniques,” Informatica, vol. 31, pp. 249-268, 2007.
- Cufoglu, A., Lohi, M., and Madani, K. “Classification accuracy performance of Naive Bayesian (NB), Bayesian Networks (BN), Lazy Learning of Bayesian Rules (LBR) and Instance-Based Learner (IB1)-comparative study,” IEEE International Conference on Computer Engineering & Systems, pp. 210-215, 2008.
- Jaber Karimpour,Ali A. Noroozi and Adeleh Abadi. “The Impact of Feature Selection on Web Spam Detection,” I.J. Intelligent Systems and Applications, vol. 4, pp. 61-67, 2012.


