New mutation of the TP53 gene associated with the hereditary breast cancer in a young tuvinian woman
Автор: Gervas Polina A., Molokov Aleksey Yu., Zarubin Aleksei A., Ponomareva Anastasia A., Babyshkina Nataliya N., Belyavskaya Valentina A., Pisareva Lubov F., Choynzonov Evgeny L., Cherdyntseva Nadezda V.
Журнал: Сибирский онкологический журнал @siboncoj
Рубрика: Случай из клинической практики
Статья в выпуске: 6 т.20, 2021 года.
Бесплатный доступ
Background. The identification of the ethnospecific mutations associated with hereditary breast cancer remains challenging. Next generation sequencing (NGS) technology fully enables the compilation of germline variants associated with the risk for inherited diseases. Despite the success of the NGS, up to 20 % of molecular tests report genetic variant of unknown significance (VUS) or novel variants that have never been previously described and their clinical significances are unknown. To obtain extended information about the variants of the unknown significance, it is necessary to use an alternative approach for the analysis of the NGS data. To obtain extended characteristic about the unknown significance variants, it is necessary to search for additional tools for the analysis of the NGS data. Material and Methods. We reclassified the mutation of the unknown significance using the ActiveDriveDB database that assessed the effect of mutations on sites of post-translational modifications, and the ProteinPaint tool that complemented the existing cancer genome portals and provided a comprehensive and intuitive view of cancer genomic data. Results. in this study, we report a 44-year-old Tuvinian woman with a family history of breast cancer. Based on the NGS data, mutational analysis revealed the presence of the LRG_321t1: c.80C>T heterozygous variant in exon 2, which led to the proline to leucine change at codon 27 of the protein. in the dbPubMed database, this mutation was determined as unknown significance due to data limitation. According to the data of the ActiveDriverDB tool, this mutation is located distally at the site of post-translational protein modification, which is responsible for binding to kinases that regulate genes of the cell cycle, etc. (ATM, CHEK2, CDK, MAPK). in accordance with ProteinPaint tool, the LRG_321t1: c.80C>T mutation is located in functionally specialized transactivation domains and codon of the TP53 gene, where the pathogenic mutation associated with Li-Fraumeni syndrome has been earlier described. Conclusion. This report is the first to describe a new variant in the TP53 gene (rs1555526933), which is likely to be associated with hereditary cancer-predisposing syndrome, including Li-Fraumeni syndrome, in a Tuvinian BC patient with young-onset and familial BC.
Germline mutation, breast cancer, small nationality of Russia, ethnic group, tuvinian
Короткий адрес: https://sciup.org/140261348
IDR: 140261348 | УДК: 618.19-006.6-056.7:575.224 | DOI: 10.21294/1814-4861-2021-20-6-164-170
Текст научной статьи New mutation of the TP53 gene associated with the hereditary breast cancer in a young tuvinian woman
Breast cancer (BC) is the most common female malignancy worldwide. Germline alterations are responsible for early-onset BC or ovarian cancer in 7–15 % of patients [1]. Next generation sequencing allows the detection of different types of DNA damage (not pathogenic, likely not pathogenic, uncertain, likely pathogenic, pathogenic). The interpretation of variants of unknown significance (VUS) is still a major challenge and misunderstanding of them can lead to serious clinical mistakes for patients and their families [2, 3]. Breast cancer-related gene alterations are known to differ between different ethnic groups. There are limited data on hereditary BC-associated mutations in Tuvinian BC patients (0,5 % of the total population of Russia) [4].
Our previous study was aimed at identifying the BC-associated mutations in 24 BC patients living in Siberia (Tuvans) using NGS. Thus, 75 % of patients with early-onset BC and/or family history had no clinically relevant mutations (pathogenic mutations). This study aimed to reclassify the variants of unknown significance by using the comprehensive human proteo-genomics ActiveDriveDB database and ProteinPaint tool that allow visualization of mutations in protein.
Material and Methods
The eligibility criteria were as follows: early age of onset BC and/or family history (2 or more close relatives with BC) and/or the presence of synchronous or metachronous multiple primary tumors [5]. Exclusion criteria included the presence of well-known BC-associated BRCA gene mutations (BRCA1 5382insC, 185delAG, 4153delAG, T300G, 3819delGTAAA, 3875delGTCT, 2080delA and BRCA2 6174delT).
Blood was collected in blood tubes containing K2EDTA. Genomic DNA was extracted from the peripheral blood lymphocytes using the phenol/ chloroform method. DNA library were prepared using the Hereditary Cancer SolutionTM kit (Sophia GENETICS, Switzerland) to cover 27 genes, such as ATM, APC, BARD1, BRCA1, BRCA2, BRIP1, CDH1, CHEK2, EPCAM, FAM175A, MLH1, MRE11A, MSH2, MSH6, MUTYH, NBN, PALB2, PIK3CA, PMS2, PMS2CL, PTEN, RAD50, RAD51C, RAD51D, STK11, TP53 , and XRCC2 . Paired-end sequencing (2 × 150 bp) was conducted using NextSeq 500 system (Illumina, USA) [6].
Bioinformatics analysis
Sequencing data was analyzed according to the GATK (Genome Analysis Toolkit) best practice recommendation for Whole Exome Sequencing using GRCh37 as a reference for Burrows-Wheeler alignment. The obtained variants were annotated with ANNOVAR software and ranged according to population frequency (genomic exome, gnomAD genome, and ExAC), ClinVar, CADD, and literature data [7–9]. Detected sequence variants were annotated using PolyPhen2, Mutation Taster, and SIFT [10–12].
The open-source database is available at (Ontario Institute for Cancer Research) and contains more than 385,000 mutations associated with post-translational modification sites (PTM mutations) and numerous amino acid substitutions from The Cancer Genome
Atlas, ClinVar and other projects and simulates site-specific interaction networks of proteins with upstream enzymes and approved drugs. The VCF files were aligned to hg19/GRCh37 version of the human genome. NGS data were further annotated regarding their position in post-translational modifications sites. Depending on the location of amino acid substitutions in the post-translational modification sites, mutations can be described as direct (central amino acid residue of the PTM site) or indirect (proximal or distal if they replace 1–2 or 3–7 amino acid residues from the nearest PTM site, respectively). An assessment of the network impact of mutations also was carried out using ActiveDriverDB. ProteinPaint tool was used to complement existing cancer genome portals and provide a comprehensive and intuitive view of cancer genomic data with advanced visualization features.
Results and Discussion
We report the case of a 44-year-old Tuvinian woman with breast cancer in early age and family history (second relatives with stomach cancer). Based on the NGS data, mutational analysis revealed the presence of the c.80C>T heterozygous variant in exon 2 of TP53 gene.
First mention about this variant was reported by S. Kato et al. (2003) [13]. This variant of TP53 gene was included to International Agency for Research on Cancer (IARC) TP53 database. The p.P27L variant (also known as c.80C>T), located in coding exon 2 of the TP53 gene, results from a C to T substitution at nucleotide position 80. The proline at codon 27 is replaced by leucine, an amino acid with similar properties. This variant is reported to have deficient transactivation capacity compared to wild type in yeast based functional assays. In silico prediction for this alteration is inconclusive. The authors summarized that clinical significance of this alteration remains unclear.
Data from PubMed and ClinVare database
The mutation NM_001126112.2: c.80C>T (HGVS) of TP53 gene is marked as rs1555526933, position chr17:7676398 (GRCh38.p13), variation type is SNV, frequency – none. In ClinVar this variant is reported as missense variant of unknown or uncertain clinical significance.
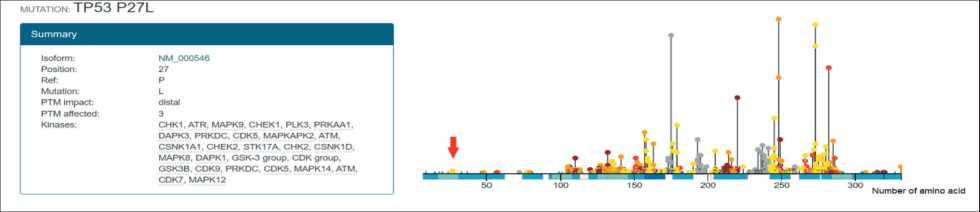
Fig. 1. Location of the mutation NM_001126112.2: c.80C>T (HGVS) of TP53 gene in the post-translational modification site (red arrow) Рис. 1. Локализация мутации NM_001126112.2: c.80C>T (HGVS) в гене TP53 в сайте посттрансляционной модификации (красная стрелка)
Data from human proteo-genomics ActiveDrive database
Open-source database https://www.ActiveDriverDB. org (Ontario Institute for Cancer Research) annotates mutations through the prism of post-translational modification sites (PTMs). It is well known that after translation proteins undergo different post-translational modifications (PTMs) or chemical modifications (phosphorylation, glycosylation, ubiquitination, SUMOylation, acetylation, and lipidation) that impact their activity and function. PTMs influence many processes in the cell, including protein activation and degradation, protein-protein interactions, chromatin organization, development, and signaling pathways associated with different types of cancers [14]. Mutations at PTM sites could alter sequence motifs linked by PTM enzymes and can affect signaling networks. Further, PTMs provide potential sites of intervention and could be used in precision cancer therapies [15].
The mutation NM_001126112.2: c.80C>T (HGVS) of TP53 gene is classified by ActiveDriverDB as post-translational modification mutations with distal replace, meaning 1–2 or 3–7 amino acid residues from the PTM site (fig. 1). Moreover, the post-translational modification site, where this mutation is located, involves in regulation of numerous genes such as CHEK1/2, CDK, MAPK, ATM and others. ActiveDriverDB analysis also suggests that this substitution of TP53 gene may induce gains of phosphosites of the four pharmaceutically targetable kinases.
Data from ProteinPaint tool
The mutation of TP53 gene that was found in a young Tuvinian breast cancer woman (44-year-old) was located in codon of gene, where the pathogenic mutation associated with Li-Fraumeni syndrome was earlier described (NM_000546.6
(TP53): c.77_80delinsAAGAACGT (p.Leu26fs)). In accordance with db PubMed, the mutation (NM_000546.6 (TP53): c.77_80delinsAAGAACGT(p. Leu26fs)) of TP53 gene is marked as rs397516438, chr17:7676398-7676401 position (GRCh38.p13), frameshift variant type and none frequency. In ClinVar, this variant is reported as likely-pathogenic for Li-Fraumeni syndrome and pathogenic for hereditary cancer-predisposing syndrome [16, 17]. The p53 gene mutations described above are located in functionally specialized transactivation domain that impacts the transcription of different genes [18].
Li-Fraumeni syndrome belongs to a group of rare inherited autosomal-dominant disorder or hereditary diseases (Hereditary cancer-predisposing syndrome) and is characterized by high genetic and phenotypic heterogeneity. Li-Fraumeni syndrome is caused by an inherited (germline) pathogenic variant of the TP53 tumor suppressor gene on chromosome 17. In patients affected by Li-Fraumeni syndrome, more than 597 germline mutations in the TP53 gene (TP53 Database (IARC)) have been described. Cancers associated with Li-Fraumeni syndrome often occur at earlier ages; 80 % of bone and soft-tissue sarcomas and breast cancer associated with the syndrome occur prior to age 45 [19]. It is known about more than 1000 families with Li-Fraumeni syndrome from 172 countries. According to the literature, there is no evidence of ethnic or geographic disparity in the occurrence of Li-Fraumeni syndrome, but a high prevalence of it has been reported in Brazil. The Southern population of Brazil with Li-Fraumeni syndrome has been associated with a specific variant of the TP53 (R337H) [20]. The population of Southern Brazil with this «founder mutation» has roughly a 60 % lifetime risk of cancers with favorable survival rates.
There were some limitations in our study. Clinical-morphological characteristics of the patient and samples of her relatives were not available for the analysis, since the material was collected more than
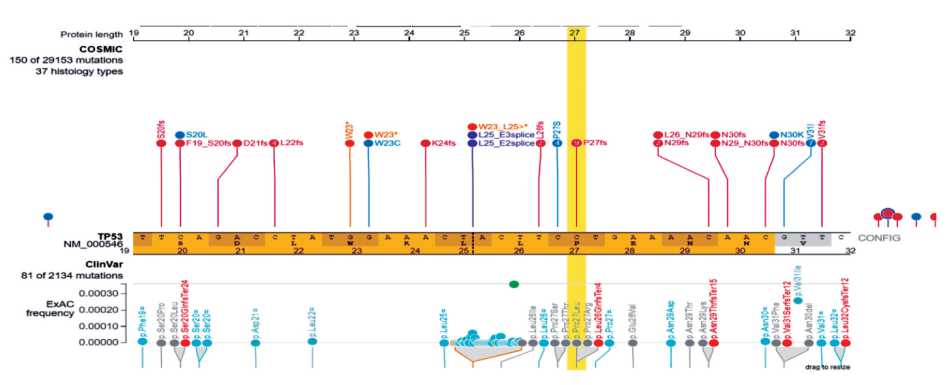
Fig. 2. Visualization of the mutation NM_001126112.2: c.80C>T (HGVS) of TP53 gene by ProteinPaint tool
Рис. 2. Визуализация мутации NM_001126112.2: c.80C>T (HGVS) в гене TP53 с помощью программы ProteinPaint
10 years ago and was partially lost. Thus, the use of additional databases (human proteo-genomics ActiveDriveDB and ProteinPaint) made it possible to obtain additional important information about NM_001126112.2: c.80C>T mutation of the TP53 gene.
Conclusion
This report is the first to describe the new variant in the TP53 gene (rs1555526933) that likely can be associated with Hereditary cancer-predisposing syndrome, including the Li-Fraumeni syndrome, in a Tuvinian BC patient with young-onset and familial BC.
Список литературы New mutation of the TP53 gene associated with the hereditary breast cancer in a young tuvinian woman
- Sokolenko A.P., Preobrazhenskaya E.V., Aleksakhina S.N., Iyevleva A.G., Mitiushkina N.V., Zaitseva O.A., Yatsuk O.S., Tiurin V.I., Strelkova T.N., Togo A.V., Imyanitov E.N. Candidate gene analysis of BRCA1/2 mutationnegative high-risk Russian breast cancer patients. Cancer Lett. 2015; 359(2): 259–61. doi: 10.1016/j.canlet.2015.01.022.
- Plon S.E., Eccles D.M., Easton D., Foulkes W.D., Genuardi M., Greenblatt M.S., Hogervorst F.B., Hoogerbrugge N., Spurdle A.B., Tavtigian S.V.; IARC Unclassified Genetic Variants Working Group. Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum Mutat. 2008 Nov; 29(11): 1282–91. doi: 10.1002/humu.20880. PMID: 18951446; PMCID: PMC3075918.
- Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., Voelkerding K., Rehm H.L.; ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015 May; 17(5): 405–24. doi: 10.1038/gim.2015.30.
- Cherdyntseva N.V., Pisareva L.F., Ivanova A.A., Panferova Y.V., Malinovskaya E.A., Odintsova I.N., Doroshenko A.V., Gervas P.A., Slonimskaya E.M., Shivit-ool A.A., Dvornichenko V.V., Choinzonov Y.L. Ethnic aspects of hereditary breast cancer in the region of Siberia. Vestn Ross Akad Med Nauk. 2014; 11–12: 72–9. doi: 10.15690/vramn.v69i11-12.1186. [Article in Russian].
- Eccles D.M. Hereditary cancer: guidelines in clinical practice. Breast and ovarian cancer genetics. Ann Oncol. 2004; 15 Suppl 4: iv133-8. doi: 10.1093/annonc/mdh917.
- Slatko B.E., Gardner A.F., Ausubel F.M. Overview of Next-Generation Sequencing Technologies. Curr Protoc Mol Biol. 2018; 122(1): e59. doi: 10.1002/cpmb.59.
- Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., Del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J., Banks E., Garimella K.V., Altshuler D., Gabriel S., DePristo M.A. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013; 43(1110): 11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43.
- DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M., McKenna A., Fennell T.J., Kernytsky A.M., Sivachenko A.Y., Cibulskis K., Gabriel S.B., Altshuler D., Daly M.J. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011 May; 43(5): 491–8. doi: 10.1038/ng.806.
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010 Sep; 20(9): 1297–303. doi: 10.1101/gr.107524.110.
- Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat Methods. 2010 Apr; 7(4): 248–9. doi: 10.1038/nmeth0410-248.
- Schwarz J.M., Cooper D.N., Schuelke M., Seelow D. Mutation-Taster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014 Apr; 11(4): 361–2. doi: 10.1038/nmeth.2890.
- Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nature protocols. 2009; 4(7): 1073–81. doi.org/10.1038/nprot.2009.86.
- Kato S., Han S.Y., Liu W., Otsuka K., Shibata H., Kanamaru R., Ishioka C. Understanding the function-structure and function-mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc Natl Acad Sci USA. 2003; 100(14): 8424–9. doi.org/10.1073/pnas.1431692100.
- Samaržija I. Post-Translational Modifications That Drive Prostate Cancer Progression. Biomolecules. 2021; 11(2): 247. doi.org/10.3390/biom11020247.
- Krassowski M., Paczkowska M., Cullion K., Huang T., Dzneladze I., Ouellette B., Yamada J.T., Fradet-Turcotte A., Reimand J. ActiveDriverDB: human disease mutations and genome variation in post-translational modification sites of proteins. Nucl Acids Res. 2018; 46(D1): D901–D910. doi.org/10.1093/nar/gkx973.
- Murakami I., Fujiwara Y., Yamaoka N., Hiyama K., Ishioka S., Yamakido M. Detection of p53 gene mutations in cytopathology and biopsy specimens from patients with lung cancer. Am J Resp Crit Care Med. 1996; 154(4 Pt 1): 1117–23. doi.org/10.1164/ajrccm.154.4.8887616.
- Heide I., Thiede C., Sonntag T., de Kant E., Neubauer A., Jonas S., Peter F.J., Neuhaus P., Herrmann R., Huhn D., Rochlitz C.F. The status of p53 in the metastatic progression of colorectal cancer. Eur J Cancer. 1997; 33(8): 1314–22. doi.org/10.1016/s0959-8049(97)00118-4.
- Sullivan K.D., Galbraith M.D., Andrysik Z., Espinosa J.M. Mechanisms of transcriptional regulation by p53. Cell Death Diff. 2018; 25(1): 133–43. doi.org/10.1038/cdd.2017.174.
- Yurgelun M.B., Masciari S., Joshi V.A., Mercado R.C., Lindor N.M., Gallinger S., Hopper J.L., Jenkins M.A., Buchanan D.D., Newcomb P.A., Potter J.D., Haile R.W., Kucherlapati R., Syngal S.; Colon Cancer Family Registry. Germline TP53 Mutations in Patients With Early-Onset Colorectal Cancer in the Colon Cancer Family Registry. JAMA Oncol. 2015 May; 1(2): 214–21. doi: 10.1001/jamaoncol.2015.0197.
- Palmero E.I., Schüler-Faccini L., Caleffi M., Achatz M.I., Olivier M., Martel-Planche G., Marcel V., Aguiar E., Giacomazzi J., Ewald I.P., Giugliani R., Hainaut P., Ashton-Prolla P. Detection of R337H, a germline TP53 mutation predisposing to multiple cancers, in asymptomatic women participating in a breast cancer screening program in Southern Brazil. Cancer Lett. 2008 Mar 8; 261(1): 21–5. doi: 10.1016/j.canlet.2007.10.044.


