Transcriptome of metastatic colorectal cancer
Автор: Dronova T.A., Babyshkina N.N., Kostromitsky D.N., Eremin D.A., Cherdyntseva N.V.
Журнал: Cardiometry @cardiometry
Статья в выпуске: 33, 2024 года.
Бесплатный доступ
Colorectal cancer has been one of the leading malignant neoplasms in males and females over many years. The vast majority of patients (70%) with colon lesions develop distant metastases to the liver, which are the main cause of death.
Colorectal cancer, liver metastases, whole-transcriptome sequencing, gene panel
Короткий адрес: https://sciup.org/148329778
IDR: 148329778 | DOI: 10.18137/cardiometry.2024.33.conf.12
Текст статьи Transcriptome of metastatic colorectal cancer
Introduction. Colorectal cancer has been one of the leading malignant neoplasms in males and females over many years [1]. The vast majority of patients (70%) with colon lesions develop distant metastases to the liver, which are the main cause of death [2]. It has been shown that the overall 5-year survival rate of such patients is 12% only [3]. Research in recent years has significantly expanded our understanding of the molecular genetic basis of colorectal cancer carcinogenesis; however, information on transcripts involved in the mechanisms of metastasis is still fragmentary [4, 5].
The aim hereof is to conduct a comparative analysis of the transcriptome of primary colon tumors and liver metastases before the stage of antitumor treatment and determine a panel of genes involved in the mechanisms of metastasis.
PATIENTS/MATERIALS AND METHODS
This study used biopsy samples of tumor and adjacent normal colon tissue, as well as metastatic nodules in the liver, obtained from 5 patients before their antitumor treatment (FOLFOXIRI chemotherapy + targeted therapy). The criteria for inclusion of patients in the study were as follows: T3-4N1-3M1; metastatic liver disease; patients’ condition according to the WHO-ECOG system 0-1 score; average age 40-70 years. The exclusion criteria were as listed below: refusal of treatment; rectal cancer with damage to the middle and lower ampullar regions. The biopsy samples were placed in RNA stabilizer (Biolabmix, Russia), incubated for 24 hours at +2–8ºС and stored at –80ºС. RNA was isolated using the PureLink RNA Mini Kit (Invi-trogen, USA). The integrity of the obtained RNA was assessed with the 4150 Tape Station (Agilent, USA). The quality and the concentration of RNA were determined with the Qubit 4.0 fluorimeter (Thermo Fisher Scientific). The concentration of the samples was 35–280 ng/μl. Libraries for sequencing were prepared using the KAPA RNA HyperPrep Kit with RiboErase (HMR) (KAPA, South Africa) according to the manufacturer’s protocol. The library sizes were from 200 to 800 bp. The analyzed samples were sequenced with the NextSeq500 device (Illumina, USA) in single-end reading mode (75 cycles). At the next stage of the research work, a bioinformatics analysis of the obtained data was performed. The quality of sequencing was assessed using the FastQC and MultiQC softwares. The alignment of reads to the reference genome was carried out with the STAR software. The counting of reads mapped to individual genes was performed using the HTSeq-count software package. Differential expression data were obtained utilizing DESeq2 (R environment).
RESULTS
At the first stage of the work, the transcriptome of primary colon tumors and liver metastases obtained from 5 patients before their treatment was analyzed. The cluster analysis of colon tumor tissue showed differential expression of 62 genes. The top 10 genes by log2FC (double logarithm of the magnitude of differences in average expression between the study groups) and padj (the probability value of differences adjusted for multiple comparisons) were as follows: CCR4, FER1L4, AMH, RHOV, GALT, CELSR3, DRAM1, RHPN1, PKMYT1, and PCSK9. Our analysis of the results of the cluster analysis of the transcriptome of liver metastases allowed revealing of 301 transcripts, including the top 10 genes as given below: MUC3A, CDCA7, SATB2, XPNPEP2, EPCAM, SCNN1A, TONSL, AIFM3, ENTPD8, and GAL3ST2.
At the next stage of the work, the functional characteristics of the detected transcripts were analyzed. It has been found that the majority of hyperexpressed genes (CCR4, RHPN1, DNM1, TMEM132A, CDH3, MELTF, ARHGAP39, SLC17A1, SCNN1A, GAL3ST2, SPIRE2) in the primary tumor and the metastases are involved in the processes of migration, adhesion, and cellular transport. The products of the RHOV, TJP3,
EPPK1, IQANK1, and FER1L4 genes are responsible for the processes of organization of the cytoplasmic membranes of cells and the cytoskeleton. The PAB-PC1L gene is involved in the processes of regulation of the mRNA stability; SATB2 and TONSL are involved in the processes of transcription regulation; and ENTPD8 is involved in the regulation of cellular metabolism. The CELSR3 and DRAM1 transcripts are associated with oncogenesis; the MMP11 transcript is necessary for the degradation of the intracellular matrix. In addition, it has been revealed that the products of the genes GRM8, FAM83E, WNK2, AIFM3, PTK6, and CDCA7 contribute to the processes of intracellular transduction, and the genes AMH and EPCAM are responsible for the regulation of cell differentiation. Transcripts DGAT1, ANO9, MUC3A, XPNPEP2 are involved in the mechanisms of metastasis and participate in the metabolism of proteins and lipids. Sepa- rately, it is worth to highlight the hyperexpressed genes in the primary colon tumor: GALT, PCSK9, ALG8, the functional significance of which is associated with the processes of metabolism of fatty acids and glucose.
At the last stage of our study, an analysis of the sharpness of biological processes was carried out based on transcripts hyperexpressed in the primary colon tumor and metastatic foci in the liver. The Enrichr database was applied for the analysis. It has been established that the metabolic processes are activated in tumor tissue before treatment as listed below: carbohydrate, protein and lipoprotein metabolism, as well as intracellular transduction processes (see Figure 1A). On the contrary, the following biological processes contribute to the implementation of metastasis: the regulation of proliferation, nucleotide metabolism, and cell adhesion (see Figure 1B).
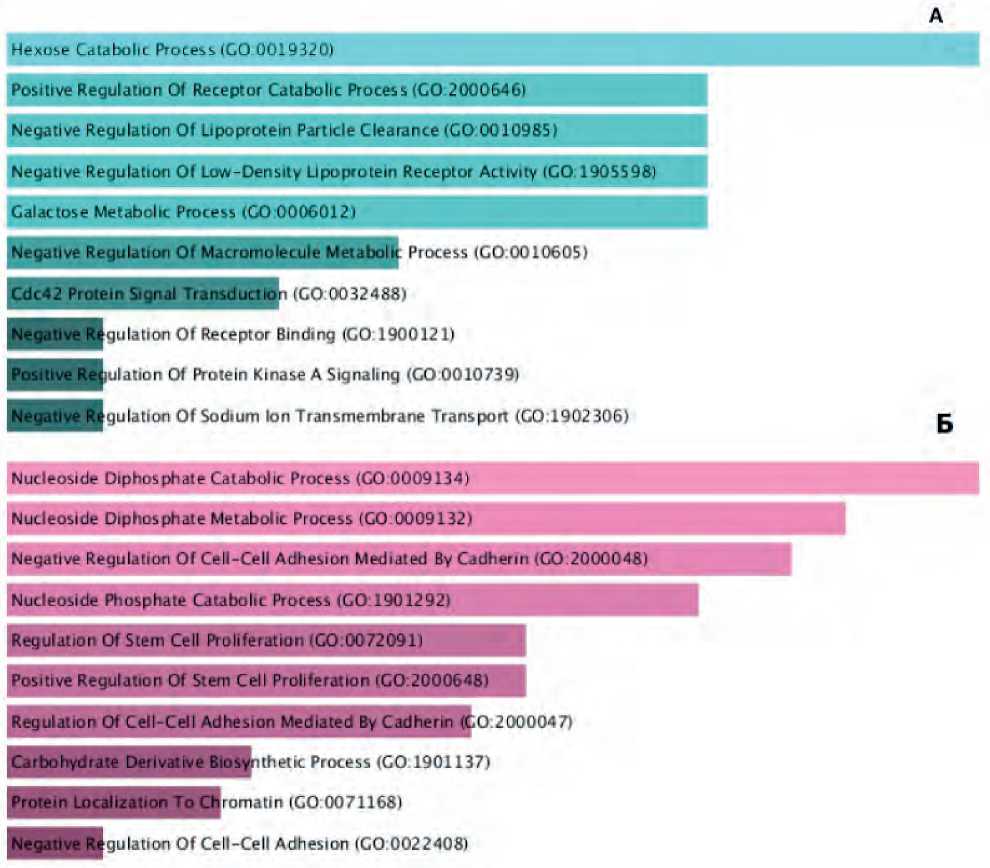
Figure 1. Biological processes expressed in the primary colon tumor (A) and liver metastases (B) before the stage of antitumor treatment, potentially involved in the processes of metastasis (p<0.05).
CONCLUSION
In the course of the research work, clusters of genes involved in the processes of metastasis of primarily disseminated colon tumors were identified. It was shown that dysregulation of proliferative intracellular programs may be of decisive importance in the mechanisms of metastasis. The processes of formation of metastasis of colorectal cancer can lead to disordering in such processes as cellular adhesion, as well as dys-regulation of intracellular transduction.
The research work has been conducted with grant financing by the Russian Research Fund, grant No. 2215-00212,
Список литературы Transcriptome of metastatic colorectal cancer
- Шахзадова А. О., Старинский В. В., Лисичникова И. В. Состояние онкологической помощи населению России в 2022 году. Сибирский онкологический журнал. 2023; 22(5): 5-13. DOI: 10.21294/1814-4861-2023-22-5-5-13
- Zarour LR, Anand S, Billingsley KG, et al. Colorectal cancer liver metastasis: evolving paradigms and future directions. Cell Mol Gastroenterol Hepatol. 2017; 3:163-73. DOI: 10.1016/j.jcmgh.2017.01.006
- Brouwer NPM, Bos A, Lemmens V, et al. An overview of 25 years of incidence, treatment and outcome of colorectal cancer patients. Int J Cancer. 2018; 143: 2758-66. DOI: 10.1002/ijc.31785
- Karaca C, Demir Karaman E, Leblebici A, et al. New treatment alternatives for primary and metastatic colorectal cancer by an integrated transcriptome and network analyses. Sci Rep. 2024; 14:8762. DOI: 10.1038/s41598-024-59101-8
- Wang R, Li J, Zhou X, et al. Single-cell genomic and transcriptomic landscapes of primary and metastatic colorectal cancer tumors. Genome Med. 2022; 14: 93. DOI: 10.1186/s13073-022-01093-z


